mTORC1 cooperates with tRNA wobble modification to sustain the protein synthesis machinery
- PMID: 40328729
- PMCID: PMC12056009
- DOI: 10.1038/s41467-025-59185-4
mTORC1 cooperates with tRNA wobble modification to sustain the protein synthesis machinery
Abstract
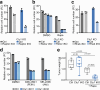
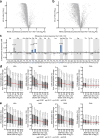
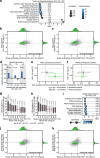
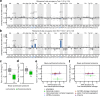
Synthesizing the cellular proteome is a demanding process that is regulated by numerous signaling pathways and RNA modifications. How precisely these mechanisms control the protein synthesis machinery to generate specific proteome subsets remains unclear. Here, through genome-wide CRISPR screens we identify genes that enable mammalian cells to adapt to inactivation of the kinase mechanistic target of rapamycin complex 1 (mTORC1), the central driver of protein synthesis. When mTORC1 is inactive, enzymes that modify tRNAs at wobble uridines (U34-enzymes), Elongator and Ctu1/2, become critically essential for cell growth in vitro and in tumors. By integrating quantitative nascent proteomics, steady-state proteomics and ribosome profiling, we demonstrate that the loss of U34-enzymes particularly impairs the synthesis of ribosomal proteins. However, when mTORC1 is active, this biosynthetic defect only mildly affects steady-state protein abundance. By contrast, simultaneous suppression of mTORC1 and U34-enzymes depletes cells of ribosomal proteins, globally inhibiting translation. Thus, mTORC1 cooperates with tRNA U34-enzymes to sustain the protein synthesis machinery and support the high translational requirements of cell growth.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.Z. is a founder, shareholder and scientific advisor of Quantro Therapeutics. J.Z. and the Zuber lab receive research support and funding from Boehringer Ingelheim. Other authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

