HiBC: a publicly available collection of bacterial strains isolated from the human gut
- PMID: 40328737
- PMCID: PMC12056005
- DOI: 10.1038/s41467-025-59229-9
HiBC: a publicly available collection of bacterial strains isolated from the human gut
Abstract
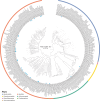
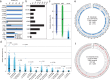
Numerous bacteria in the human gut microbiome remain unknown and/or have yet to be cultured. While collections of human gut bacteria have been published, few strains are accessible to the scientific community. We have therefore created a publicly available collection of bacterial strains isolated from the human gut. The Human intestinal Bacteria Collection (HiBC) ( https://www.hibc.rwth-aachen.de ) contains 340 strains representing 198 species within 29 families and 7 phyla, of which 29 previously unknown species are taxonomically described and named. These included two butyrate-producing species of Faecalibacterium and new dominant species associated with health and inflammatory bowel disease, Ruminococcoides intestinale and Blautia intestinihominis, respectively. Plasmids were prolific within the HiBC isolates, with almost half (46%) of strains containing plasmids, with a maximum of six within a strain. This included a broadly occurring plasmid (pBAC) that exists in three diverse forms across Bacteroidales species. Megaplasmids were identified within two strains, the pMMCAT megaplasmid is globally present within multiple Bacteroidales species. This collection of easily searchable and publicly available gut bacterial isolates will facilitate functional studies of the gut microbiome.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Groussin, M. et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell184, 2053–2067.e18 (2021). - PubMed
MeSH terms
Grants and funding
- 513892404/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 445552570/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 395357507/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 403224013/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 460129525/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources

