Genome-wide association study of post COVID-19 syndrome in a population-based cohort in Germany
- PMID: 40328884
- PMCID: PMC12056214
- DOI: 10.1038/s41598-025-00945-z
Genome-wide association study of post COVID-19 syndrome in a population-based cohort in Germany
Abstract
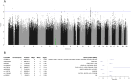
If health impairments due to coronavirus disease 2019 (COVID-19) persist for 12 weeks or longer, patients are diagnosed with Post-COVID Syndrome (PCS), or Long-COVID. Although the COVID-19 pandemic has largely subsided in 2024, PCS is still a major health burden worldwide, and identifying potential genetic modifiers of PCS remains of great clinical and scientific interest. We therefore performed a case-control type genome-wide association study (GWAS) of three recently developed PCS (severity) scores in 2,247 participants of COVIDOM, a prospective, multi-centre, population-based cohort study of SARS-CoV-2-infected individuals in Germany. Each PCS score originally represented the weighted sum of the binary indicators of all, or a subset, of 12 PCS symptom complexes, assessed six months or later after the PCR test-confirmed SARS-CoV-2 infection of a participant. For various methodical reasons, however, the PCS scores were dichotomized along their respective median values in the present study, prior to the GWAS. Of the 6,383,167 single nucleotide polymorphisms included, various variants were found to be associated with at least one of the PCS scores, although not at the stringent genome-wide statistical significance level of 5 × 10- 8. With p = 6.6 × 10- 8, however, the genotype-phenotype association of SNP rs9792535 at position chr9:127,166,653 narrowly missed this threshold. The SNP is located in a region including the NEK6, PSMB7 and ADGRD2 genes which, however, does not immediately suggest an etiological connection to PCS. As regards functional plausibility, variants of a possible effect mapped to the olfactory receptor gene region (lead SNP rs10893121 at position chr11:123,854,744; p = 2.5 × 10- 6). Impairment of smell and taste is a pathognomonic feature of both, acute COVID-19 and PCS, and our results suggest that this connection may have a genetic basis. Three other genotype-phenotype associations pointed towards a possible etiological role in PCS of cellular virus repression (CHD6 gene region), activation of macrophages (SLC7A2) and the release of virus particles from infected cells (ARHGAP44). All other gene regions highlighted by our GWAS did not relate to pathophysiological processes currently discussed for PCS. Therefore, and because the genotype-phenotype associations observed in our GWAS were generally not very strong, the complexity of the genetic background of PCS appears to be as high as that of most other multifactorial traits in humans.
Keywords: Genotype-phenotype association; Linkage disequilibrium; Long-COVID; Macrophage activation; Olfactory receptor; SARS-CoV-2; Single nucleotide polymorphism; Virus repression.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

