Influence of orthodontic appliances and nitrate on the oral microbiota
- PMID: 40328933
- PMCID: PMC12055954
- DOI: 10.1007/s00253-025-13496-0
Influence of orthodontic appliances and nitrate on the oral microbiota
Abstract
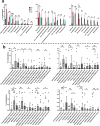
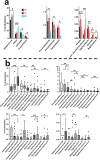
In this pilot study, we investigated the bacterial changes introduced on the subgingival, tongue, and saliva microbiota during fixed orthodontic treatment, with or without daily administration of nitrate-containing beet juice for 2 weeks in 22 individuals with good general health. We followed clinical parameters in combination with microbiota changes before, after 2 weeks, and after 6 months of treatment with fixed orthodontic appliances. In accordance with variations in community composition at the sampling sites, effects to orthodontic treatment differed. Subgingival communities responded promptly to orthodontic treatment with no additional structural changes over time, whereas saliva and tongue communities were affected only after extended treatment. Periodontal pathogens such as Selenomonas sputigena were enriched in subgingival communities, whereas Streptococcus mutans was enriched in saliva. Specifically, Rothia mucilaginosa increased tremendously in relative abundance in both tongue and saliva communities. The effect of beet juice on microbial composition was significant in subgingival samples even though the differences were not mirrored in single differentially distributed genera or species. This indicates changes in the complete subgingival microbial net of interacting species. However, the prevention of Corynebacterium matruchotii enrichment by beet juice may be important for prevention of biofilm formation. Enrichment of Neisseria flavescens group bacteria and Abiotrophia and depletion of different Actinomyces and Stomatobaculum were observed on tongue communities. We conclude that subgingival microbiota are rapidly affected by fixed orthodontic appliances and can be positively influenced by regular administration of nitrate-containing juice. KEY POINTS: • The subgingival site, tongue, and saliva contain different microbiota • The microbiota react differently to orthodontic treatment and beet juice • Key genera and species affected by treatments were identified.
Keywords: Dietary supplements; Gingival disease; Nitrate; Oral health; Oral microbiota; Orthodontic fixed appliances.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: The study was approved by the Medical Ethics Committees of the University of Wuerzburg, Germany, and Northwest and Central Switzerland (trial register: 185–2017; 2019–01896). All parents/legal guardians of the participants signed informed consent before participation. Conflict of interest: The authors declare no competing interests.
Figures





References
-
- Alexander SA (1991) Effects of orthodontic attachments on the gingival health of permanent second molars. Am J Orthod Dentofacial Orthop 100:337–340. 10.1016/0889-5406(91)70071-4 - PubMed
-
- Babikow E, Ghaltakhchyan N, Livingston T, Qu Y, Liu C, Hoxie A, Sulkowski T, Bocklage C, Marsh A, Phillips ST, Mitchell KB, Ribeiro AA, Jackson TH, Roach J, Wu D, Divaris K, Jacox LA (2024) Longitudinal microbiome changes in supragingival biofilm transcriptomes induced by orthodontics. JDR Clin Trans Res 9:265–276. 10.1177/23800844231199393 - PMC - PubMed
-
- Camarinha-Silva A, Jauregui R, Chaves-Moreno D, Oxley AP, Schaumburg F, Becker K, Wos-Oxley ML, Pieper DH (2014) Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol 16:2939–2952. 10.1111/1462-2920.12362 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

