Genomic insights into the spread of methicillin-resistant Staphylococcus aureus involved in ear infections
- PMID: 40329191
- PMCID: PMC12054198
- DOI: 10.1186/s12879-025-11052-9
Genomic insights into the spread of methicillin-resistant Staphylococcus aureus involved in ear infections
Abstract
Background: Methicillin-resistant Staphylococcus aureus (MRSA) is a major pathogen causing ear infections. However, genomic epidemiology and determinants influencing transmission of ear infections associated MRSA (EIA-MRSA) in community remain unknown.
Methods: In 2020-2021, 105 EIA-MRSA isolates were collected and sequenced from outpatients across different households in Shanghai, China. Antimicrobial susceptibility testing, core genome MLST, and phylodynamic analyses were conducted to characterize EIA-MRSA dissemination.
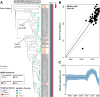
Results: Quinolone resistance was identified as a risk factor for EIA-MRSA spread (OR 9, [95% CI 3-31]). The ST764 clone and two subclones of ST22-PT hypervirulent clone have developed an extensive quinolone-resistant (eQR) phenotype, conferring additional resistance to advanced quinolones due to the accumulation of four mutations in gyrA (S84L and either S85P, E88K, or E88G) and parC (S80F and either E84K or E84G). These ST764- and ST22-PT-eQR isolates were highly transmissible and showed increased resistance to other commonly used antimicrobials, posing potential high-risk clones. The eQR phenotype may be inherent to the ST764 lineage, which emerged in the late 1980s, coinciding with the widespread fluoroquinolone usage. The ST22-PT-eQR subclones emerged in around 2017 and are accumulating resistance genes.
Conclusion: Vigilance is crucial for eQR high-risk clones, particularly the convergent ST22-PT-eQR subclones that accumulate resistance and virulence traits, posing risks for ear infections.
Clinical trial number: Not applicable.
Keywords: Antimicrobial resistance; Ear infections; Methicillin-resistant staphylococcus aureus; Transmission; Whole genome sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study conformed to the Declaration of Helsinki and was approved by the Institutional Review Boards of Eye & ENT Hospital (number: EENT2015011). Informed consent was obtained from all individual participants included in the study. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Shangali A, Kamori D, Massawe W, Masoud S, Kibwana U, Mwingwa AG, Manisha A, Mwandigha AM, Mirambo MM, Mshana SE, et al. Aetiology of ear infection and antimicrobial susceptibility pattern among patients attending otorhinolaryngology clinic at a tertiary hospital in Dar Es Salaam, Tanzania: a hospital-based cross-sectional study. BMJ Open. 2023;13(4):e068359. - PMC - PubMed
-
- Chua K, Laurent F, Coombs G, Grayson ML, Howden BP. Antimicrobial resistance: not community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA)! A clinician’s guide to community MRSA - its evolving antimicrobial resistance and implications for therapy. Clin Infect Dis. 2011;52(1):99–114. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

