In utero human intestine contains maternally derived bacterial metabolites
- PMID: 40329366
- PMCID: PMC12054239
- DOI: 10.1186/s40168-025-02110-0
In utero human intestine contains maternally derived bacterial metabolites
Abstract
Background: Understanding when host-microbiome interactions are first established is crucial for comprehending normal development and identifying disease prevention strategies. Furthermore, bacterially derived metabolites play critical roles in shaping the intestinal immune system. Recent studies have demonstrated that memory T cells infiltrate human intestinal tissue early in the second trimester, suggesting that microbial components such as peptides that can prime adaptive immunity and metabolites that can influence the development and function of the immune system are also present in utero. Our previous study reported a unique fetal intestinal metabolomic profile with an abundance of several bacterially derived metabolites and aryl hydrocarbon receptor (AHR) ligands implicated in mucosal immune regulation.
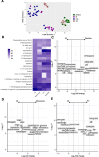
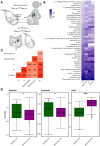
Results: In the current study, we demonstrate that a number of microbiome-associated metabolites present in the fetal intestines are also present in the placental tissue, and their abundance is different across the fetal intestine, fetal meconium, fetal placental villi, and the maternal decidua. The fetal gastrointestinal samples and maternal decidua samples show substantially higher positive correlation on the abundance of these microbial metabolites than the correlation between the fetal gastrointestinal samples and meconium samples. The expression of genes associated with the transport and signaling of some microbial metabolites is also detectable in utero.
Conclusions: We suggest that the microbiome-associated metabolites are maternally derived and vertically transmitted to the fetus. Notably, these bacterially derived metabolites, particularly short-chain fatty acids and secondary bile acids, are likely biologically active and functional in regulating the fetal immune system and preparing the gastrointestinal tract for postnatal microbial encounters, as the transcripts for their various receptors and carrier proteins are present in second trimester intestinal tissue through single-cell transcriptomic data. Video Abstract.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Placental and fetal samples were obtained from the University of Pittsburgh Biospecimen Core from electively terminated products of conception (14–23 weeks of gestation) with IRB approval and signed informed consent (IRB no. 18010491, University of Pittsburgh). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Update of
-
In utero human intestine contains maternally derived bacterial metabolites.bioRxiv [Preprint]. 2024 Aug 21:2024.08.20.608888. doi: 10.1101/2024.08.20.608888. bioRxiv. 2024. Update in: Microbiome. 2025 May 6;13(1):116. doi: 10.1186/s40168-025-02110-0. PMID: 39229010 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

