Multi-omics analysis reveals immunosuppression in oesophageal squamous cell carcinoma induced by creatine accumulation and HK3 deficiency
- PMID: 40329370
- PMCID: PMC12057037
- DOI: 10.1186/s13073-025-01465-1
Multi-omics analysis reveals immunosuppression in oesophageal squamous cell carcinoma induced by creatine accumulation and HK3 deficiency
Abstract
Background: Deep insights into the metabolic remodelling effects on the immune microenvironment of oesophageal squamous cell carcinoma (ESCC) are crucial for advancing precision immunotherapies and targeted therapies. This study aimed to provide novel insights into the molecular landscape of ESCC and identify clinically actionable targets associated with immunosuppression driven by metabolic changes.
Methods: We performed metabolomic and proteomic analyses combined with previous genomic and transcriptomic data, identified multi-omics-linked molecular features, and constructed metabolic-immune interaction-based ESCC classifiers in a discovery cohort and an independent validation cohort. We further verified the molecular characteristics and related mechanisms of ESCC subtypes.
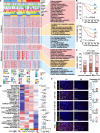
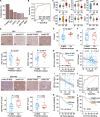
Results: Our integrated multi-omics analysis revealed dysregulated proteins and metabolic imbalances characterizing ESCC, with significant alterations in metabolites and proteins linked to genetic traits. Importantly, ESCC patients were stratified into three subtypes (S1, S2, and S3) on the basis of integrated metabolomic and proteomic data. A robust subtype prediction model was developed and validated across two independent cohorts. Notably, patients classified under the poorest prognosis subtype (S3 subtype) exhibited a significant immunosuppressive microenvironment. We identified key metabolism-related biomarkers for the S3 subtype, specifically creatine and hexokinase 3 (HK3). Creatine accumulation and HK3 protein deficiency synergistically reprogrammed macrophage metabolism, driving M2-like TAM polarization. This metabolic shift fostered an immunosuppressive microenvironment that accelerated tumour progression. These results highlight the potential of targeting creatine metabolism to improve the efficacy of immunotherapy and targeted therapy for ESCC.
Conclusions: Our analysis reveals molecular variation in multi-omics linkages and identifies targets that reverse the immunosuppressive microenvironment through metabolic remodelling improving immunotherapy and targeted therapy for ESCC.
Keywords: Creatine; Hexokinase 3; Immunosuppressive microenvironment; Metabolomics; Multi-omics; Oesophageal squamous cell carcinoma; Proteomics; Subtypes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by Shanxi Medical University's ethical committees. Informed consent was obtained from all participants. The research adhered to the declaration of Helsinki (2016LL106, 2013103). The animal experiments were approved by the Ethics Committee of Shanxi Medical University (SYDL2023040). All animal work was conducted complying with the Guidance of the Shanxi Medical University Institutional Animal Care and Use Committee. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel R, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–63. - PubMed
-
- Jing H, Jianming X, Yun C, Wu Z, Yiping Z, Zhendong C, et al. Camrelizumab versus investigator’s choice of chemotherapy as second-line therapy for advanced or metastatic oesophageal squamous cell carcinoma (ESCORT): a multicentre, randomised, open-label, phase 3 study. Lancet Oncol. 2020;21(6):832–42. - PubMed
-
- Niu F, Yu Y, Li Z, Ren Y, Li Z, Ye Q, et al. Arginase: An emerging and promising therapeutic target for cancer treatment. Biomedicine Pharmacother. 2022;149:112840. - PubMed
MeSH terms
Substances
Grants and funding
- 82302916/National Natural Science Foundation of China
- 82103143/National Natural Science Foundation of China
- 82172930/National Natural Science Foundation of China
- 82394411/National Natural Science Foundation of China
- U21A20372/National Natural Science Foundation of China
- 2021YFC2501001/National Key Research and Development Program of China
- C2303002/the Shenzhen Medical Research Fund
- C2303002/the Shenzhen Medical Research Fund
- ZYYC23013/the West China Hospital 135 project
- 201605D131045-16/the Special Fund for Science and Technology Innovation Teams of Shanxi Province
- 202204051001024/the Science and Technology Innovation Team of Shanxi Province
- 202204021301062/the Science and Technology Achievements Transformation Project of Shanxi Province
- BYJL005/Shanxi Province Higher Education "Billion Project" Science and Technology Guidance Project
LinkOut - more resources
Full Text Sources
Medical

