Integrating large-scale meta-GWAS and PigGTEx resources to decipher the genetic basis of 232 complex traits in pigs
- PMID: 40330097
- PMCID: PMC12051865
- DOI: 10.1093/nsr/nwaf048
Integrating large-scale meta-GWAS and PigGTEx resources to decipher the genetic basis of 232 complex traits in pigs
Abstract
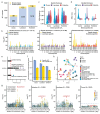
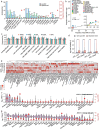
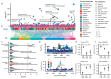
Understanding the molecular and cellular mechanisms underlying complex traits in pigs is crucial for enhancing genetic gain via artificial selection and utilizing pigs as models for human disease and biology. Here, we conducted comprehensive genome-wide association studies (GWAS) followed by a cross-breed meta-analysis for 232 complex traits and a within-breed meta-analysis for 12 traits, using 28.3 million imputed sequence variants in 70 328 animals across 14 pig breeds. We identified 6878 quantitative trait loci (QTL) for 139 complex traits. Leveraging the Pig Genotype-Tissue Expression resource, we systematically investigated the biological context and regulatory mechanisms behind these trait-QTLs, ultimately prioritizing 14 829 variant-gene-tissue-trait regulatory circuits. For instance, rs344053754 regulates UGT2B31 expression in the liver and intestines, potentially by modulating enhancer activity, ultimately influencing litter weight at weaning in pigs. Furthermore, we observed conservation of certain genetic and regulatory mechanisms underlying complex traits between humans and pigs. Overall, our cross-breed meta-GWAS in pigs provides invaluable resources and novel insights into the genetic regulatory and evolutionary mechanisms of complex traits in mammals.
Keywords: PigGTEx; complex traits; meta-GWAS; molQTL; pig.
© The Author(s) 2025. Published by Oxford University Press on behalf of China Science Publishing & Media Ltd.
Figures



References
-
- FAO. 2023 . Meat Market Review: Emerging Trends and Outlook. 2023. https://openknowledge.fao.org/handle/20.500.14283/cd0465en
LinkOut - more resources
Full Text Sources
