Genome-wide identification of the nuclear redox protein gene family revealed its potential role in drought stress tolerance in rice
- PMID: 40330127
- PMCID: PMC12052764
- DOI: 10.3389/fpls.2025.1562718
Genome-wide identification of the nuclear redox protein gene family revealed its potential role in drought stress tolerance in rice
Abstract
Introduction: Thioredoxins (TRX) are redox-active proteins critical for plant stress adaptation. As a TRX family member, nucleoredoxin (NRX) maintains drought-induced redox homeostasis, yet its genome-wide characterization in rice remains uninvestigated.
Methods: Using HMMER3.0 (E-value <1e-5) and TBtools, we identified OsNRX genes across three rice varieties (Minghui63, Nipponbare, 9311). Conserved domains were verified by SMART/CDD, while promoter cis-elements were systematically predicted with PlantCARE. Tissue-specific expression patterns were analyzed using RiceXPro data, and drought responses were quantified via qRT-PCR in drought-tolerant (Jiangnong Zao 1B) versus sensitive (TAISEN GLUTINOUS YU 1157) varieties under PEG6000 stress.
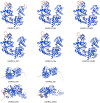
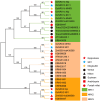
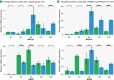
Results: Ten OsNRX genes were classified into three subfamilies (NRX1/NRX2/NRX3) exhibiting conserved domain architectures. Promoter analysis identified abundant stress-responsive elements (ABRE, MBS) and phytohormone signals (ABA/JA/SA). Tissue-specific expression profiles revealed NRX1a dominance in roots/hulls, versus NRX1b/NRX2 enrichment in endosperm. Drought stress triggered rapid OsNRX upregulation (20-53-fold within 3-6h), with tolerant varieties showing earlier NRX2 activation than sensitive counterparts.
Discussion: OsNRX genes exhibit dynamic drought-responsive regulation, while their spatiotemporal expression in glumes, embryos, and endosperm suggests potential dual roles in stress adaptation and grain development. These results provide molecular targets for improving drought resilience in rice breeding.
Keywords: drought stress; expression pattern; genome-wide identification; nucleoredoxin gene family; rice.
Copyright © 2025 Liu, Zheng, Yang, Li and Yin.
Conflict of interest statement
Author HY was employed by Huazhi Biotechnology Co. Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Calderón A., Sevilla F., Jiménez A. (2018). “Redox protein thioredoxins: function under salinity, drought and extreme temperature conditions,” in Antioxidants and antioxidant enzymes in higher plants. Eds. Gupta D. K., Palma J. M., Corpas F. J. (Springer International Publishing, Cham: ), 123–162. doi: 10.1007/978-3-319-75088-0_7 - DOI
-
- Cha J.-Y., Ahn G., Jeong S. Y., Shin G.-I., Ali I., Ji M. G., et al. (2022). Nucleoredoxin 1 positively regulates heat stress tolerance by enhancing the transcription of antioxidants and heat-shock proteins in tomato. Biochem. Biophys. Res. Commun. 635, 12–18. doi: 10.1016/j.bbrc.2022.10.033 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

