Genetic diversity and population structure of Anatolian Hair goats, an ancient breed
- PMID: 40330641
- PMCID: PMC12051132
- DOI: 10.5194/aab-67-13-2024
Genetic diversity and population structure of Anatolian Hair goats, an ancient breed
Abstract
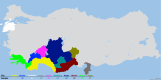
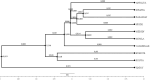
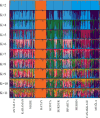
This study aimed to investigate the genetic characterization and diversity of Hair goats from 10 regions using 20 microsatellite markers. A total of 522 alleles were observed. The INRA0023 locus exhibited the greatest number of alleles (48), while the DRBP1 locus had the highest effective allele number (16.27), and the BM1818 and DRBP1 loci had the highest polymorphic information content value (0.94). The expected heterozygosity value ranged from 0.85 (ILSTS011) to 0.94 (BM1818, SRCRSP15, and DRBP1). The Hair goat populations in Konya and Hatay displayed the lowest and highest allele numbers, with values of 10.40 and 16.25, respectively. The fixation index ( ) values are significant in defining population structures and determining the extent of heterozygosity losses. The values exhibited a range of 0.031 in Muǧla to 0.226 in Burdur. A total of 107 unique alleles were identified in Hair goat populations. However, it is noteworthy that, out of all the alleles, only 25 had a frequency exceeding 5 %. The results indicate that the microsatellite markers utilized demonstrate sufficient levels of polymorphism, making them appropriate for efficiently investigating the genetic variability of Hair goat populations.
Copyright: © 2024 Aylin Demiray et al.
Conflict of interest statement
The contact author has declared that none of the authors has any competing interests.
Figures

References
-
- Agaoglu OK, Ertugrul O. Assessment of genetic diversity, genetic relationship and bottleneck using microsatellites in some native Turkish goat breeds. Small Rumin Res. 2012;105:53–60. doi: 10.1016/j.smallrumres.2011.12.005. - DOI
-
- Al-Atiyat RM, Alobre MM, Aljumaah RS, Alshaikh MA. Microsatellite based genetic diversity and population structure of three Saudi goat breeds. Small Rumin Res. 2015;130:90–94. doi: 10.1016/j.smallrumres.2015.07.027. - DOI
-
- Anonymous [last access: 15 March 2023]. 2023. https://www.esk.gov.tr/tr/11124/Kil-Kecisi-Kara-Keci.
-
- Atay O, Gökdal Ö, Eren V. Reproductive characteristics and kid marketing weights of hair goat flocks in rural conditions in Türkiye. Cuban J Agr Sci. 2010;44:353–358.
-
- Awobajo OK, Salako AE, Osaiyuwu OH. Analysis of genetic structure of Nigerian West African Dwarf goats by microsatellite markers. Small Rumin Res. 2015;133:112–117.
LinkOut - more resources
Full Text Sources
