This is a preprint.
DNA2 and MSH2 activity collectively mediate chemically stabilized G4 for efficient telomere replication
- PMID: 40330857
- PMCID: PMC12051496
- DOI: 10.1101/2025.04.04.647332
DNA2 and MSH2 activity collectively mediate chemically stabilized G4 for efficient telomere replication
Abstract
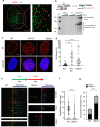
G-quadruplexes (G4s) are widely existing stable DNA secondary structures in mammalian cells. A long-standing hypothesis is that timely resolution of G4s is needed for efficient and faithful DNA replication. In vitro, G4s may be unwound by helicases or alternatively resolved via DNA2 nuclease mediated G4 cleavage. However, little is known about the biological significance and regulatory mechanism of the DNA2-mediated G4 removal pathway. Here, we report that DNA2 deficiency or its chemical inhibition leads to a significant accumulation of G4s and stalled replication forks at telomeres, which is demonstrated by a high-resolution technology: Single molecular analysis of replicating DNA (SMARD). We further identify that the DNA repair complex MutSα (MSH2-MSH6) binds G4s and stimulates G4 resolution via DNA2-mediated G4 excision. MSH2 deficiency, like DNA2 deficiency or inhibition, causes G4 accumulation and defective telomere replication. Meanwhile, G4-stabilizing environmental compounds block G4 unwinding by helicases but not G4 cleavage by DNA2. Consequently, G4 stabilizers impair telomere replication and cause telomere instabilities, especially in cells deficient in DNA2 or MSH2.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures





References
-
- Waga S. & Stillman B. The DNA replication fork in eukaryotic cells. Annual review of biochemistry 67, 721–751 (1998). - PubMed
-
- Lopes M. et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412, 557–561 (2001). - PubMed
-
- Bell S.P. & Dutta A. DNA replication in eukaryotic cells. Annual review of biochemistry 71, 333–374 (2002). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
