The role of Tim-3+T cell subsets in the peripheral blood of patients with COVID-19 and diabetes
- PMID: 40330891
- PMCID: PMC12053769
- DOI: 10.1016/j.isci.2025.112339
The role of Tim-3+T cell subsets in the peripheral blood of patients with COVID-19 and diabetes
Abstract
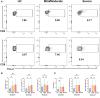
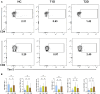
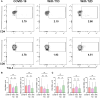
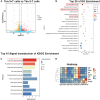
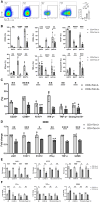
Corona Virus Disease 2019 (COVID-19) and diabetes interact to influence disease severity, yet their combined immunological characteristics remain unclear. Here, we analyzed Tim-3+ T cells in patients with COVID-19, Type 1 Diabetes (T1D), or both conditions. COVID-19 reduced peripheral T cell subsets but increased Tim-3+ cells, while T1D and COVID-19 with T1D showed the opposite pattern. Patients with Type 2 Diabetes (T2D) exhibited no significant alterations. In human samples and mouse models, Tim-3+ T cells demonstrated impaired activation and cytokine production. RNA-seq analysis in mice and RT-PCR analysis in human samples together identified the dysregulation of the JAK-STAT pathway in Tim-3+ T cells. These findings highlight Tim-3-mediated JAK-STAT dysregulation in T-cells as a potential mechanism linking COVID-19 and T1D, offering insights for therapeutic targeting.
Keywords: Disease; Immune response; Immunology.
© 2025 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Altered Tim-1 and IL-10 Expression in Regulatory B Cell Subsets in Type 1 Diabetes.Front Immunol. 2021 Dec 21;12:773896. doi: 10.3389/fimmu.2021.773896. eCollection 2021. Front Immunol. 2021. PMID: 35754999 Free PMC article.
-
Altered expression of Tim family molecules and an imbalanced ratio of Tim-3 to Tim-1 expression in patients with type 1 diabetes.Front Endocrinol (Lausanne). 2022 Jul 28;13:937109. doi: 10.3389/fendo.2022.937109. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35966054 Free PMC article.
-
Ebola Virus Binding to Tim-1 on T Lymphocytes Induces a Cytokine Storm.mBio. 2017 Sep 26;8(5):e00845-17. doi: 10.1128/mBio.00845-17. mBio. 2017. PMID: 28951472 Free PMC article.
-
Role of Tim-3 in COVID-19: a potential biomarker and therapeutic target.Arch Virol. 2023 Jul 31;168(8):213. doi: 10.1007/s00705-023-05842-2. Arch Virol. 2023. PMID: 37522944 Review.
-
T-cell immunoglobulin and mucin-domain containing-3 (TIM-3): Solving a key puzzle in autoimmune diseases.Int Immunopharmacol. 2023 Aug;121:110418. doi: 10.1016/j.intimp.2023.110418. Epub 2023 Jun 6. Int Immunopharmacol. 2023. PMID: 37290326 Review.
References
LinkOut - more resources
Full Text Sources
Research Materials

