Antiviral defense against filovirus infections: targets and evasion mechanisms
- PMID: 40331244
- PMCID: PMC12153392
- DOI: 10.1080/17460913.2025.2501924
Antiviral defense against filovirus infections: targets and evasion mechanisms
Abstract
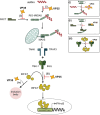
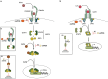
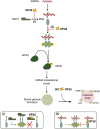
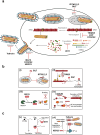
Filoviruses include a number of serious human pathogens, infections with which result in the development of hemorrhagic fevers with high case fatality rates. As for other RNA viruses, viral replication generates both protein and RNA species that can serve as danger signals, leading to the activation of antiviral defense pathways. However, in order to be able to efficiently infect humans these viruses have developed mechanisms that allow them to evade diverse host antiviral defense mechanisms. Consequently, in addition to their functions within the viral lifecycle many filovirus proteins have been shown to have accessory functions involved in the regulation of diverse host pathways. These include those of the type-I interferon response, other pathways involved in dsRNA-sensing, as well as the selective inhibition of interferon stimulated gene activities. Further, filoviruses have developed mechanisms to subvert recognition of infected cells and the generation of neutralizing antibodies. This review focuses on bringing together the evidence to date supporting the existence of diverse mechanisms aimed at regulating these pathways as well as providing details of the mechanisms involved.
Keywords: Ebola virus; Filoviruses; adaptive immunity; immune antagonism; immune evasion; innate immunity; virus-host cell interactions.
Plain language summary
Filoviruses like Ebola virus and Marburg virus can cause serious disease when they infect humans. During the infection these viruses generate danger signals that can be detected by the immune system to trigger defense pathways that can help combat these infections. Therefore, to be able to successfully infect humans, these viruses have needed to develop various ways to block these defense pathways. Many filovirus proteins have been shown to have these kinds of activities. Some of the mechanisms involved are common to many different filoviruses, but others are specific for certain filoviruses. Also, while different filoviruses may block the same pathway, they sometimes do so using different virus proteins or different mechanisms. This review summarizes our current knowledge about how filoviruses interact with host defense pathways to achieve successful infection and also how this can play a role in the development of disease.
Conflict of interest statement
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Figures
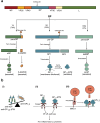
References
-
- Bodmer BS, Breithaupt A, Heung M, et al. In vivo characterization of the novel ebolavirus Bombali virus suggests a low pathogenic potential for humans. Emerg Microbes Infect. 2023;12(1):2164216. doi: 10.1080/22221751.2022.2164216 - DOI - PMC - PubMed
-
• First assessment of a novel filovirus regarding its pathogenic potential.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
