[Exosome separation and analysis based on microfluidics technology and its clinical applications]
- PMID: 40331610
- PMCID: PMC12059995
- DOI: 10.3724/SP.J.1123.2024.10032
[Exosome separation and analysis based on microfluidics technology and its clinical applications]
Abstract
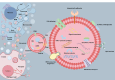
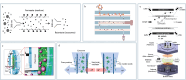
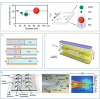
Exosomes are cell-secreted nanoscale vesicles 30-150 nm in size and encompass a diverse array of biomolecules, including lipids, proteins, and nucleic acids. Exosomes play pivotal roles during the intercellular exchange of materials and information, and are closely associated with the onset and progression of a variety of diseases. Therefore, comprehensively investigating exosomes is very important in terms of disease diagnosis and treatment. However, exosomes are genetically heterogeneous and are composed of different materials. Additionally, exosome-size and packing-specific-biomarker heterogeneities result in biofunction diversity. Moreover, isolating and analyzing exosomes is highly challenging owing to their small sizes and heterogeneities. Accordingly, effective separation methods and analytical techniques for highly specifically and efficiently identifying exosomes are urgently needed in order to better understand their functionalities. While separation and analysis is required to reveal exosome heterogeneity, the former is confronted by three primary challenges. Firstly, exosome heterogeneity (including heterogeneous marker expressions and size heterogeneity that results in heterogeneous functions) results in systems that are very difficult to separate. Secondly, the coexistence of non-vesicular contaminants (lipoprotein nanoparticles, soluble proteins, nucleic acids, etc.) and the complex matrix effects of body fluids also contribute to separation difficulties. Thirdly, enrichment is a highly challenging task owing to low exosome concentrations. Traditional methods, such as ultracentrifugation and size-exclusion chromatography, fall short in terms of their abilities to precisely separate and analyze exosomes. On the other hand, microfluidics has emerged as a robust tool for the efficient analysis of complex biological samples and is characterized by miniaturization, precise control, high throughput, automation, and integration. Firstly, the operability, integrability, and modifiability of a microfluidics system facilitate exosome separation and purification based on surface properties, size, charge, and polarity. Secondly, the use of a microfluidics approach, with its high throughput, low reagent consumption, and multichannel manipulability, greatly facilitates preparing exosomes and enhancing their concentrations. Thirdly, microfluidics ensures that diverse separation methods are compatible with downstream analysis techniques. Exosomes are highly heterogeneous; hence, they are classified by type and subpopulation (according to origin, size, molecular markers, functions, etc.). This paper first discusses microfluidics techniques for separating exosomes and examines various separation strategies grounded in the physicochemical properties of exosomes. We then analyze exosome detection methodologies that use microfluidics platforms and encompass traditional group-exosome analysis techniques and novel single-exosome analysis approaches. Finally, we discuss future clinical applications of microfluidics technology in exosome research, particularly its potential for diagnosing and treating diseases, thereby underscoring the applications value of microfluidics technology in the realm of personalized and precision medicine. Furthermore, cutting-edge microfluidics platforms offer novel perspectives for purifying and preparing EVs owing to precise fluid control, integration, miniaturization, and high-throughput characterization. EV populations, subpopulations, and single vesicles can be purified based on their physicochemical properties and microfluidics features. Comprehensive lab-on-a-chip methods are promising in terms of separating EVs based on traits, such as size, surface markers, and charge, and for obtaining highly pure EVs. Recycled EV samples can be prepared by controlling the high-throughput and multichannel capabilities of microfluidics approaches. The transition from bulk EV analysis to single-vesicle analysis provides opportunities to explore the heterogeneous nature of EVs, thereby augmenting their potential for disease diagnosis.
外泌体是由细胞分泌的纳米级囊泡,其中富含脂质、蛋白质、核酸等多种生物功能分子。外泌体在生物体内的信息传递、疾病机理研究及诊断等方面扮演着类似“物联网、互联网”的关键角色。然而,受限于外泌体来源、尺寸、内含物以及功能的异质性,外泌体分离分析仍然面临诸多挑战。传统外泌体分离分析方法包括超速离心、尺寸排阻和免疫共沉淀等,但这些方法普遍存在产量低、纯度低等问题限制了其进一步的临床应用。微流控技术因其微型化、高通量、自动化和集成化等特点为复杂生物样本的高效分离分析提供了强有力的工具。基于此,本文系统总结了基于微流控技术的外泌体分离和分析方法。首先,讨论了基于外泌体的尺寸、电荷、表面官能团等物化性质结合微流控技术分离外泌体及外泌体亚群的分离方法;其次,分析了基于微流控平台的外泌体检测手段,包括外泌体群体分析方法和单个外泌体分析方法;最后,探讨了微流控技术在外泌体临床应用中的前景,并在疾病诊断和治疗方面进行了展望。微流控技术有望突破传统研究中的分离分析瓶颈,为癌症等重大疾病的精准诊断及治疗提供新技术、新平台。
Keywords: disease diagnosis and treatment; exosomes; microfluidics; separation analysis.
Figures
References
-
- Cocozza F, Grisard E, Martin-Jaular L, et al. . Cell, 2020, 182(1): 262 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous