Characterization of the family-level Borreliaceae pan-genome and development of an episomal typing protocol
- PMID: 40331826
- PMCID: PMC12153284
- DOI: 10.1128/mbio.00943-25
Characterization of the family-level Borreliaceae pan-genome and development of an episomal typing protocol
Abstract
The Borreliaceae family includes many obligate parasitic bacterial species etiologically associated with a myriad of zoonotic borrelioses, including Lyme disease and vector-borne relapsing fevers. Borreliaceae infections are difficult to detect by both direct and indirect methods, often leading to delayed and missed diagnoses. Efforts to improve diagnostics center around the development of molecular diagnostics (MDx), but due to deep tissue sequestration and the lack of persistent bacteremias, even MDx assays suffer from a lack of sensitivity. Additionally, the extensive genomic heterogeneity among isolates, even within the same species, contributes to the lack of assay sensitivity, as single target assays, whether nucleic acid-based or serologically based, cannot provide universal coverage. This within-species heterogeneity is partly due to differences in replicon repertoires and genomic structures that have likely arisen to support the complex Borreliaceae life cycle necessary for these parasites to survive in multiple hosts, each with unique immune responses. We constructed a Borreliaceae family-level pan-genome and characterized the phylogenetic relationships among the constituent taxa, which supports the recent, although contested, taxonomy of splitting the family into at least two genera. Gene content profiles were created for the majority of the Borreliaceae replicons, providing for the first time their unambiguous molecular typing. Our characterization of the Borreliaceae pan-genome supports the splitting of the former Borrelia genus into two genera and provides for the phylogenetic placement of several non-species designated isolates. Mining this family-level pan-genome will enable the development of precision diagnostics corresponding to gene content-driven clinical outcomes while also providing targets for interventions.
Importance: Using whole genome sequencing, we demonstrated that the bacteria that are transmitted by ticks and other arthropod vectors that cause Lyme disease and relapsing fevers, while related, do not belong within the same genus classification. In addition, through characterization of their highly atypical genomic structure, we were able to develop a genetic typing system that will help with future studies of how they cause disease while also providing targets for medical interventions.
Keywords: Borrelia; Borreliaceae; Borreliella; comparative genomics; distributed genome hypothesis; pan-genome; phylogenetics; spirochete; supragenome; taxonomy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



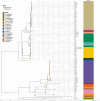
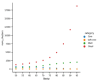


Update of
-
Characterization of the family-level Borreliaceae pan-genome and development of an episomal typing protocol.Res Sq [Preprint]. 2024 Jun 11:rs.3.rs-4491589. doi: 10.21203/rs.3.rs-4491589/v1. Res Sq. 2024. Update in: mBio. 2025 Jun 11;16(6):e0094325. doi: 10.1128/mbio.00943-25. PMID: 38947078 Free PMC article. Updated. Preprint.
References
-
- Shen K, Wang X, Post JC, Ehrlich GD. 2003. Molecular and translational research approaches for the study of bacterial pathogenesis in otitis media, p 91–119. In Rosenfeld R, Bluestone CD (ed), Evidence-based otitis media, 2nd ed. B. C. Decker Inc, Hamilton, Ontario, Canada.
-
- Ehrlich GD, Hu ZF, Post JC. 2004. Role for biofilms in infectious disease, p 332–358. In Ghannoum M, O’Toole GA (ed), Microbial biofilms. ASM Press, Washington, D.C.
-
- Ehrlich GD, Ahmed A, Earl J, Hiller NL, Costerton JW, Stoodley P, Post JC, DeMeo P, Hu FZ. 2010. The distributed genome hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol Med Microbiol 59:269–279. doi: 10.1111/j.1574-695X.2010.00704.x - DOI - PMC - PubMed
-
- Shen K, Antalis P, Gladitz J, Sayeed S, Ahmed A, Yu S, Hayes J, Johnson S, Dice B, Dopico R, Keefe R, Janto B, Chong W, Goodwin J, Wadowsky RM, Erdos G, Post JC, Ehrlich GD, Hu FZ. 2005. Identification, distribution, and expression of novel genes in 10 clinical isolates of nontypeable Haemophilus influenzae. Infect Immun 73:3479–3491. doi: 10.1128/IAI.73.6.3479-3491.2005 - DOI - PMC - PubMed
MeSH terms
Grants and funding
- AI 141801/National Institute of Allergy and Infectious Diseases
- R01 DC002148/DC/NIDCD NIH HHS/United States
- Bill an Marian Cook Foundation
- DK 082316/DK/NIDDK NIH HHS/United States
- Oskar Fischer Project
- DC 01248/DC/NIDCD NIH HHS/United States
- U01 AI169840/AI/NIAID NIH HHS/United States
- U01 DK082316/DK/NIDDK NIH HHS/United States
- R01 AI165876/AI/NIAID NIH HHS/United States
- U24 DK082316/DK/NIDDK NIH HHS/United States
- R01 AI141801/AI/NIAID NIH HHS/United States
- AI 165876/National Institute of Allergy and Infectious Diseases
LinkOut - more resources
Full Text Sources
