Cyclosporine A Accelerates Neurorecovery Transcriptional Trajectory in a Swine Model of Diffuse Traumatic Brain Injury
- PMID: 40331981
- PMCID: PMC12026708
- DOI: 10.3390/ijms26083531
Cyclosporine A Accelerates Neurorecovery Transcriptional Trajectory in a Swine Model of Diffuse Traumatic Brain Injury
Abstract
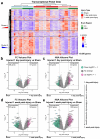
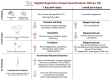
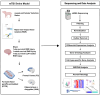
Mild traumatic brain injury (mTBI) is a leading cause of morbidity in children with both short- and long-term neurological, cognitive, cerebrovascular, and emotional deficits. These deficits have been attributed to ongoing pathophysiological cascades that occur acutely and persist post-injury. Given our limited understanding of the transcriptional changes associated with these pathophysiological cascades, we studied formalin-fixed paraffin-embedded (FFPE) tissues from the frontal cortex (FC) and the hippocampus + amygdala (H&A) regions of swine (N = 40) after a sagittal rapid non-impact head rotation (RNR). We then sequenced RNA to define transcriptional changes at 1 day and 1 week after injury and investigated the protective influence of cyclosporine A (CsA) treatment. Differentially expressed genes (DEGs) were classified into five temporal patterns (Early, Transient, Persistent, Intensified, Delayed, or Late). DEGs were more abundant at 1 week than 1 day. Shared significant gene ontology annotations in both regions included terms associated with neuronal distress at 1 day and neurorecovery at 1 week. CsA (20 mg/kg/day) infused for 1 day (beginning at 6 h after injury) accelerated 466 DEGs in the FC and 2794 DEGs in the H&A, such that the CsA-treated transcriptional profile was associated with neurorecovery. Overall, our data reveal the effects of anatomic region and elapsed time on gene expression post-mTBI and motivate future studies of CsA treatment.
Keywords: RNA-seq; TBI; gene expression; pediatric; pig; treatment; weighted gene co-expression network analysis.
Conflict of interest statement
The funding institutions had no role in the conceptualization, study design, data collection, analysis, preparation, writing, review, and decision to submit this manuscript for publication.
Figures






Similar articles
-
Analysis of miRNA Expression Profiles in Traumatic Brain Injury (TBI) and Their Correlation with Survival and Severity of Injury.Int J Mol Sci. 2024 Sep 2;25(17):9539. doi: 10.3390/ijms25179539. Int J Mol Sci. 2024. PMID: 39273487 Free PMC article.
-
Integrated Bioinformatics Analysis for the Identification of Key Molecules and Pathways in the Hippocampus of Rats After Traumatic Brain Injury.Neurochem Res. 2020 Apr;45(4):928-939. doi: 10.1007/s11064-020-02973-9. Epub 2020 Jan 30. Neurochem Res. 2020. PMID: 31997105
-
Synaptic Mitochondria Sustain More Damage than Non-Synaptic Mitochondria after Traumatic Brain Injury and Are Protected by Cyclosporine A.J Neurotrauma. 2017 Apr 1;34(7):1291-1301. doi: 10.1089/neu.2016.4628. Epub 2016 Oct 13. J Neurotrauma. 2017. PMID: 27596283 Free PMC article.
-
Identification of Key Genes and Pathways in the Hippocampus after Traumatic Brain Injury: Bioinformatics Analysis and Experimental Validation.J Integr Neurosci. 2023 Feb 20;22(2):44. doi: 10.31083/j.jin2202044. J Integr Neurosci. 2023. PMID: 36992583
-
Quantitative Proteomic Study Reveals Up-Regulation of cAMP Signaling Pathway-Related Proteins in Mild Traumatic Brain Injury.J Proteome Res. 2018 Feb 2;17(2):858-869. doi: 10.1021/acs.jproteome.7b00618. Epub 2017 Dec 27. J Proteome Res. 2018. PMID: 29215295
References
-
- Georges A., M Das J. StatPearls. StatPearls Publishing; Treasure Island, FL, USA: 2022. Traumatic Brain Injury.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

