Phenotypic and Genotypic Bacterial Virulence and Resistance Profiles in Hidradenitis Suppurativa
- PMID: 40331997
- PMCID: PMC12026681
- DOI: 10.3390/ijms26083502
Phenotypic and Genotypic Bacterial Virulence and Resistance Profiles in Hidradenitis Suppurativa
Abstract
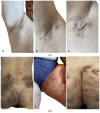
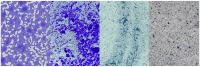
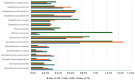
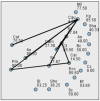
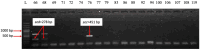
Hidradenitis suppurativa (HS) is a chronic inflammatory skin condition, primarily affecting young individuals, with a significant impact on their quality of life due to recurrent, painful nodules, abscesses, and oozing sinus tracts, primarily affecting intertriginous areas. The pathogenesis of HS is multifactorial, involving a complex interplay between genetic predisposition, immune dysregulation, microbial, and environmental factors. While it is known that cutaneous and gut microbiome contribute to innate immune dysregulation in HS, their precise involvement in disease pathogenesis remains unclear. Despite several studies investigating the microbiome of HS lesions, either by culture-dependent or independent methods, there is no data available on the interplay between bacterial virulence profiles, clinical manifestations, and the host immune response. This study aimed to explore the phenotypic and genotypic resistance and virulence profiles of microorganisms isolated from HS lesions (including the expression of soluble virulence factors and the ability to develop biofilms), with a special focus on Staphylococcus aureus (S. aureus), one of the most frequent infectious agents of HS. A total of 92 bacterial strains, belonging to 20 different bacterial species, were isolated from the HS lesions of 23 patients. The strains of Staphylococcus, Corynebacterium, and Enterococcus expressed the highest levels of soluble virulence factors, such as hemolysins, lecithinase, and lipase, which are involved in bacterial persistence, local invasivity, and tissue damage. Moreover, a significant variation among bacterial species was noted regarding the capacity to develop biofilms, with a potential impact on disease chronicization, bacterial tolerance to antibiotics, and immune defense mechanisms. The genetic characterization of methicillin-resistant staphylococci revealed the presence of adhesins, hemolysin and enterotoxin genes as well as methicillin and macrolides resistance genes. Our findings highlight the critical role of virulence determinants, including bacterial biofilms, in HS pathogenesis, emphasizing the need for targeted therapeutic strategies to disrupt biofilms and mitigate infection severity.
Keywords: Staphylococcus aureus; biofilm; hidradenitis suppurativa; skin microbiome; virulence factors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Zouboulis C.C., Bechara F.G., Fritz K., Goebeler M., Hetzer F.H., Just E., Kirsten N., Kokolakis G., Kurzen H., Nikolakis G., et al. S2k guideline for the treatment of hidradenitis suppurativa/acne inversa—Short version. J. Dtsch. Dermatol. Ges. J. Ger. Soc. Dermatol. JDDG. 2024;22:868–889. doi: 10.1111/ddg.15412. - DOI - PubMed
-
- Zouboulis C.C., Bechara F.G., Benhadou F., Bettoli V., Bukvić Mokos Z., Del Marmol V., Dolenc-Voljč M., Giamarellos-Bourboulis E.J., Grimstad Ø., Guillem P., et al. European S2k guidelines for hidradenitis suppurativa/acne inversa part 2: Treatment. J. Eur. Acad. Dermatol. Venereol. JEADV. 2024;00:1–43. doi: 10.1111/jdv.20472. - DOI - PMC - PubMed
-
- Zouboulis C.C., Desai N., Emtestam L., Hunger R.E., Ioannides D., Juhasz I., Lapins J., Matusiak L., Prens E.P., Revuz J., et al. European S1 guideline for the treatment of hidradenitis suppurativa/acne inversa. J. Eur. Acad. Dermatol. Venereol. JEADV. 2015;29:619–644. doi: 10.1111/jdv.12966. - DOI - PubMed
-
- Ingram J.R., Jenkins-Jones S., Knipe D.W., Morgan C.L.I., Cannings-John R., Piguet V. Population-based Clinical Practice Research Datalink study using algorithm modelling to identify the true burden of hidradenitis suppurativa. Br. J. Dermatol. 2018;178:917–924. doi: 10.1111/bjd.16101. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

