Mutagenesis of Intrinsically Disordered Domain Impacts Topoisomerase IIα Catalytic Activity
- PMID: 40332112
- PMCID: PMC12026768
- DOI: 10.3390/ijms26083604
Mutagenesis of Intrinsically Disordered Domain Impacts Topoisomerase IIα Catalytic Activity
Abstract
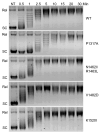
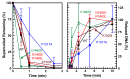
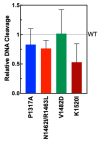
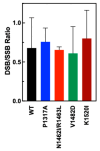
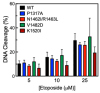
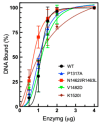
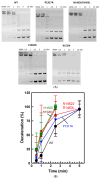
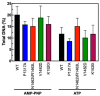
Human topoisomerase IIα and IIβ regulate DNA topology and knots in chromosomes during crucial cellular processes, making these enzymes common targets for anticancer drugs. However, selective inhibition of topoisomerase IIα (TOP2A) is desired to decrease adverse effects, which may be mediated by topoisomerase IIβ (TOP2B). The main region of difference between the two isoforms is the intrinsically disordered C-terminal domain (CTD), which is being studied as a target for selective inhibition. Our previous work examined several regions within the CTD to determine whether those regions impact biochemical function. In this current study, we designed and constructed four TOP2A mutants with amino acid substitutions in the CTD, which were then assessed for impact on biochemical activity. V1482D exhibited increased levels of relaxation, while both V1482D and K1520I exhibited increased levels of decatenation. No major impact on DNA cleavage or binding were observed with any of the mutants. The isolated impact of the changes on relaxation and decatenation supports the concept that the CTD can affect one aspect of the enzyme's function in an isolated manner, which was seen in our previous study. Taken together, these results suggest that modification of specific positions within the CTD affects substrate selection. These results are mapped onto the CTD for consideration of potential regions to target for inhibition of TOP2A.
Keywords: DNA; PSICalc; TOP2A; carboxy-terminal domain; cleavage; decatenation; intrinsically disordered domain; protein disorder; relaxation; topoisomerase IIα.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Roles of the C-terminal domains of topoisomerase IIα and topoisomerase IIβ in regulation of the decatenation checkpoint.Nucleic Acids Res. 2017 Jun 2;45(10):5995-6010. doi: 10.1093/nar/gkx325. Nucleic Acids Res. 2017. PMID: 28472494 Free PMC article.
-
Bioinformatic Analysis of Topoisomerase IIα Reveals Interdomain Interdependencies and Critical C-Terminal Domain Residues.Int J Mol Sci. 2024 May 23;25(11):5674. doi: 10.3390/ijms25115674. Int J Mol Sci. 2024. PMID: 38891861 Free PMC article.
-
Bimodal recognition of DNA geometry by human topoisomerase II alpha: preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain.Biochemistry. 2008 Dec 16;47(50):13169-78. doi: 10.1021/bi800453h. Biochemistry. 2008. PMID: 19053267 Free PMC article.
-
Topoisomerase IIα in chromosome instability and personalized cancer therapy.Oncogene. 2015 Jul 30;34(31):4019-31. doi: 10.1038/onc.2014.332. Epub 2014 Oct 20. Oncogene. 2015. PMID: 25328138 Free PMC article. Review.
-
Advances in research on malignant tumors and targeted agents for TOP2A (Review).Mol Med Rep. 2025 Feb;31(2):50. doi: 10.3892/mmr.2024.13415. Epub 2024 Dec 13. Mol Med Rep. 2025. PMID: 39670307 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

