Predicting Motif-Mediated Interactions Based on Viral Genomic Composition
- PMID: 40332242
- PMCID: PMC12028151
- DOI: 10.3390/ijms26083674
Predicting Motif-Mediated Interactions Based on Viral Genomic Composition
Abstract
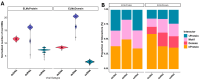
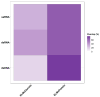
Viruses manipulate host cellular machinery to propagate their life cycle, with one key strategy being the mimicry of short linear motifs (SLiMs) found in host proteins. While databases continue to expand with virus-host protein-protein interaction (vhPPI) data, accurately predicting viral mimicry remains challenging due to the inherent degeneracy of SLiMs. In this study, we investigate how viral genomic composition influences motif mimicry and the mechanisms through which viruses hijack host cellular functions. We assessed domain-motif interaction (DMI) enrichment differences, and also predicted new DMIs based on known viral motifs with varying stringency levels, using SLiMEnrich v.1.5.1. Our findings reveal that dsDNA viruses capture significantly more known DMIs compared to other viral groups, with dsRNA viruses also exhibiting higher DMI enrichment than ssRNA viruses. Additionally, we identified new vhPPIs mediated via SLiMs, particularly within different viral genomic contexts. Understanding these interactions is vital for elucidating viral strategies to hijack host functions, which could inform the development of targeted antiviral therapies.
Keywords: bioinformatics; short linear motifs; viral mimicry; virus–host interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Prediction of motif-mediated viral mimicry through the integration of host-pathogen interactions.Arch Microbiol. 2024 Feb 9;206(3):94. doi: 10.1007/s00203-024-03832-9. Arch Microbiol. 2024. PMID: 38334822 Free PMC article.
-
Dynamic, but Not Necessarily Disordered, Human-Virus Interactions Mediated through SLiMs in Viral Proteins.Viruses. 2021 Nov 26;13(12):2369. doi: 10.3390/v13122369. Viruses. 2021. PMID: 34960638 Free PMC article. Review.
-
Virome-wide analysis of histone modification mimicry motifs carried by viral proteins.Virol Sin. 2024 Oct;39(5):793-801. doi: 10.1016/j.virs.2024.09.004. Epub 2024 Sep 16. Virol Sin. 2024. PMID: 39293541 Free PMC article.
-
Large-scale phage-based screening reveals extensive pan-viral mimicry of host short linear motifs.Nat Commun. 2023 Apr 26;14(1):2409. doi: 10.1038/s41467-023-38015-5. Nat Commun. 2023. PMID: 37100772 Free PMC article.
-
How pathogens use linear motifs to perturb host cell networks.Trends Biochem Sci. 2015 Jan;40(1):36-48. doi: 10.1016/j.tibs.2014.11.001. Epub 2014 Dec 1. Trends Biochem Sci. 2015. PMID: 25475989 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

