Identifying Genes Associated with the Anticancer Activity of a Fluorinated Chalcone in Triple-Negative Breast Cancer Cells Using Bioinformatics Tools
- PMID: 40332279
- PMCID: PMC12027753
- DOI: 10.3390/ijms26083662
Identifying Genes Associated with the Anticancer Activity of a Fluorinated Chalcone in Triple-Negative Breast Cancer Cells Using Bioinformatics Tools
Abstract
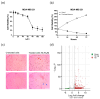
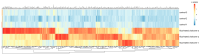
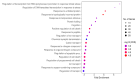
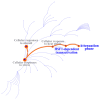
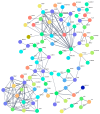
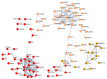
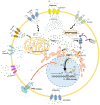
Fluorinated chalcones are molecules reported to possess potent anticancer properties against triple-negative breast cancer (TNBC) cells. However, their molecular mechanisms have not yet been fully explored. Using bioinformatics tools, we analyzed the transcriptomes of MDA-MB-231 cells treated with either a novel fluorinated chalcone (compound 3) or a control in order to identify differentially expressed (DE) genes associated with its anticancer activity and determine the biological processes in which these genes are involved. A fluorinated chalcone was synthesized using the Claisen-Schmidt method. The transcriptome of MDA-MB-231 cells was then analyzed on an Illumina NextSeq500, and DE genes with significant changes in expression were identified using the DESeq2 v1.38.0 bioinformatics tool under the strict detection criteria of |log2FC| ≥ 2 and adjusted p < 0.05. We identified 504 DE genes, which were enriched in terms related to "regulation of cell death", "cation transport", "response to topologically incorrect proteins", and "response to unfolded proteins". Surprisingly, these genes were involved in "the HSF1-dependent transactivation pathway" and "the attenuation phase pathway". This bioinformatics-based study suggests that the tested fluorinated chalcone could influence HSF-1 silencing in addition to promoting the up-regulation of several genes involved in stress-induced apoptosis. Therefore, the tested compound could have enormous potential as a novel approach for TNBC treatment.
Keywords: DE genes; TNBC; bioinformatic tools; fluorinated chalcone.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
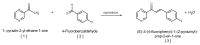
References
-
- Tavares D.F., Chaves Ribeiro V., Andrade M.A.V., Moreira Cardoso-Júnior L., Rhangel Gomes Teixeira T., Ramos Varrone G., Lopes Britto R. Immunotherapy using PD-1/PDL-1 inhibitors in metastatic triple-negative breast cancer: A systematic review. Oncol. Rev. 2021;15:497. doi: 10.4081/oncol.2021.497. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

