NPC86 Increases LncRNA Gas5 In Vivo to Improve Insulin Sensitivity and Metabolic Function in Diet-Induced Obese Diabetic Mouse Model
- PMID: 40332318
- PMCID: PMC12027414
- DOI: 10.3390/ijms26083695
NPC86 Increases LncRNA Gas5 In Vivo to Improve Insulin Sensitivity and Metabolic Function in Diet-Induced Obese Diabetic Mouse Model
Abstract
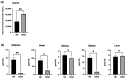
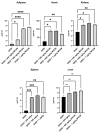
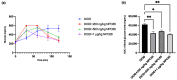
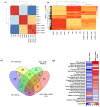
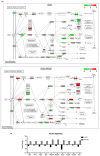
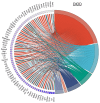
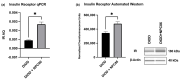
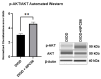
In the United States, an estimated 38 million individuals (10% of the population) have type 2 diabetes mellitus (T2D), while approximately 97.6 million adults (38%) have prediabetes. Long noncoding RNAs (lncRNAs) are critical regulators of gene expression and metabolism. We were the first to demonstrate that lncRNA Growth Arrest-Specific Transcript 5 (GAS5 (human)/gas5 (mouse)) is decreased in the serum of T2D patients and established GAS5 as a biomarker for T2D diagnosis and onset prediction, now validated by other groups. We further demonstrated that GAS5 depletion impaired glucose uptake, decreased insulin receptor levels, and inhibited insulin signaling in human adipocytes, highlighting its potential as a therapeutic target in T2D. To address this, we developed NPC86, a small-molecule compound that stabilizes GAS5 by disrupting its interaction with UPF-1, an RNA helicase involved in nonsense-mediated decay (NMD) that regulates RNA stability. NPC86 increased GAS5 and insulin receptor (IR) levels, enhanced insulin signaling, and improved glucose uptake in vitro. In this study, we tested the efficacy of NPC86 in vivo in a diet-induced obese diabetic (DIOD) mouse model, and NPC86 treatment elevated gas5 levels, improved glucose tolerance, and enhanced insulin sensitivity, with no observed toxicity or weight changes. A transcriptomics analysis of adipose tissue revealed the upregulation of insulin signaling and metabolic pathways, including oxidative phosphorylation and glycolysis, while inflammatory pathways were downregulated. These findings highlight NPC86's therapeutic potential in T2D.
Keywords: NPC86; RNA sequencing (RNAseq); diet-induced obese diabetic (DIOD); gas5; glucose metabolism; inflammation and insulin signaling; insulin resistance; long noncoding RNA (lncRNA); metabolic pathways; type 2 diabetes mellitus (T2D).
Conflict of interest statement
The authors declare no conflicts of interest regarding this manuscript. The patents are not yet licensed, and the authors have no financial conflicts of interest.
Figures





Similar articles
-
Small molecule targeting long noncoding RNA GAS5 administered intranasally improves neuronal insulin signaling and decreases neuroinflammation in an aged mouse model.Sci Rep. 2023 Jan 6;13(1):317. doi: 10.1038/s41598-022-27126-6. Sci Rep. 2023. PMID: 36609440 Free PMC article.
-
Stabilization of lncRNA GAS5 by a Small Molecule and Its Implications in Diabetic Adipocytes.Cell Chem Biol. 2019 Mar 21;26(3):319-330.e6. doi: 10.1016/j.chembiol.2018.11.012. Epub 2019 Jan 17. Cell Chem Biol. 2019. PMID: 30661991 Free PMC article.
-
Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet β-Cell Function.Cell Physiol Biochem. 2018;46(1):335-350. doi: 10.1159/000488434. Epub 2018 Mar 22. Cell Physiol Biochem. 2018. PMID: 29590649
-
Metabolism and insulin signaling in common metabolic disorders and inherited insulin resistance.Dan Med J. 2014 Jul;61(7):B4890. Dan Med J. 2014. PMID: 25123125 Review.
-
GAS5: A pivotal lncRNA in diabetes mellitus pathogenesis and management.Pathol Res Pract. 2024 Jan;253:154955. doi: 10.1016/j.prp.2023.154955. Epub 2023 Nov 19. Pathol Res Pract. 2024. PMID: 38016351 Review.
References
-
- Stratton I.M., Adler A.I., Neil H.A.W., Matthews D.R., Manley S.E., Cull C.A., Hadden D., Turner R.C., Holman R.R. Association of glycaemia with macrovascular and microvascular complications of type 2 diabetes (UKPDS 35): Prospective observational study. BMJ. 2000;321:405–412. doi: 10.1136/bmj.321.7258.405. - DOI - PMC - PubMed
-
- CDC A Report Card: Diabetes in the United States Infographic, Diabetes. [(accessed on 29 January 2025)];2024 Available online: https://www.cdc.gov/diabetes/communication-resources/diabetes-statistics....
-
- Centers for Disease Control and Prevention . National Diabetes Statistics Report, 2023: Estimates of Diabetes and Its Burden in the United States. U.S. Department of Health and Human Services; Nov, 2023. [(accessed on 8 April 2025)]. Available online: https://nationaldppcsc.cdc.gov/s/article/CDC-National-Diabetes-Statistic....
-
- Chamine I., Hwang J., Valenzuela S., Marino M., Larson A.E., Georgescu J., Latkovic-Taber M., Angier H., DeVoe J.E., Huguet N. Acute and Chronic Diabetes-Related Complications Among Patients With Diabetes Receiving Care in Community Health Centers. Diabetes Care. 2022;45:e141–e143. doi: 10.2337/dc22-0420. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

