Identification of Novel Staphylococcus aureus Core and Accessory Virulence Patterns in Chronic Rhinosinusitis
- PMID: 40332362
- PMCID: PMC12027640
- DOI: 10.3390/ijms26083711
Identification of Novel Staphylococcus aureus Core and Accessory Virulence Patterns in Chronic Rhinosinusitis
Abstract
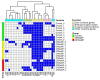
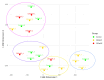
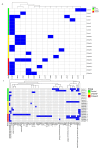
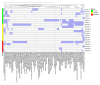
Staphylococcus aureus (S. aureus) colonizes the nasal cavities of both healthy individuals and patients with chronic rhinosinusitis (CRS) with (CRSwNP) and without (CRSsNP) nasal polyps. Treatment-resistant S. aureus biofilms and intracellular persistence are common in CRS patients, requiring the expression of specific virulence factor genes to transition into these forms. We hypothesized that S. aureus isolates from non-diseased controls, CRSsNP patients, and CRSwNP patients would exhibit distinct virulence factor patterns contributing to persistence and intracellular survival in CRS patients. Nasal swabs from seventy-seven individuals yielded S. aureus cultures in eight non-diseased controls, eight CRSsNP patients, and five CRSwNP patients. Whole-genome sequencing analyzed stress, antimicrobial resistance, and virulence genes, including plasmids and prophages. Four virulence factor gene patterns emerged: a core set (hlgA, icaC, hlgB, hlgC, hld, and aur) present in all isolates, and accessory sets, including the enterotoxin gene cluster (seo, sem, seu, sei, and sen) and a partial/complete invasive virulence factor set (splE, splA, splB, lukE, and lukD) (p = 0.001). CRSwNP isolates exhibited incomplete carriage of the core set, with frequent loss of scn, icaC, and hlgA (p < 0.05). These findings suggest that S. aureus has clusters of virulence factors that may act in concert to support the survival and persistence of the bacteria, resulting in enhanced pathogenicity. This may manifest clinically with resistant disease and refractoriness to antibiotics.
Keywords: Staphylococcus aureus; biofilms; chronic rhinosinusitis; quorum sensing; virulence factors.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Prophages encoding human immune evasion cluster genes are enriched in Staphylococcus aureus isolated from chronic rhinosinusitis patients with nasal polyps.Microb Genom. 2021 Dec;7(12):000726. doi: 10.1099/mgen.0.000726. Microb Genom. 2021. PMID: 34907894 Free PMC article.
-
[Analysis of virulence genes of Staphylococcus aureus in nasal secretionsof patients with chronic rhinosinusitis].Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2017 Sep 20;31(18):1393-1397. doi: 10.13201/j.issn.1001-1781.2017.18.004. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2017. PMID: 29797991 Chinese.
-
Detection and quantification of Staphylococcus in chronic rhinosinusitis.Int Forum Allergy Rhinol. 2019 Dec;9(12):1462-1469. doi: 10.1002/alr.22425. Epub 2019 Sep 4. Int Forum Allergy Rhinol. 2019. PMID: 31483577
-
[The role of Staphylococcus aureus in the occurrence and development of chronic rhinosinusitis with nasal polyps].Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2025 Jul;39(7):679-685. doi: 10.13201/j.issn.2096-7993.2025.07.015. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2025. PMID: 40555497 Review. Chinese.
-
Bacterial biofilms and chronic rhinosinusitis.Allergy Asthma Proc. 2013 Jul-Aug;34(4):335-341. doi: 10.2500/aap.2013.34.3665. Epub 2013 Mar 7. Allergy Asthma Proc. 2013. PMID: 23484638 Review.
References
-
- Van Zele T., Gevaert P., Watelet J.B., Claeys G., Holtappels G., Claeys C., van Cauwenberge P., Bachert C. Staphylococcus aureus colonization and IgE antibody formation to enterotoxins is increased in nasal polyposis. J. Allergy Clin. Immunol. 2004;114:981–983. doi: 10.1016/j.jaci.2004.07.013. - DOI - PubMed
-
- Maniakas A., Asmar M.H., Renteria Flores A.E., Nayan S., Alromaih S., Mfuna Endam L., Desrosiers M.Y. Staphylococcus aureus on Sinus Culture Is Associated With Recurrence of Chronic Rhinosinusitis After Endoscopic Sinus Surgery. Front. Cell Infect. Microbiol. 2018;8:150. doi: 10.3389/fcimb.2018.00150. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

