Gene Expression Profile of Cultured Human Coronary Arterial Endothelial Cells Exposed to Serum from Chronic Kidney Disease Patients: Role of MAPK Signaling Pathway
- PMID: 40332370
- PMCID: PMC12027878
- DOI: 10.3390/ijms26083732
Gene Expression Profile of Cultured Human Coronary Arterial Endothelial Cells Exposed to Serum from Chronic Kidney Disease Patients: Role of MAPK Signaling Pathway
Abstract
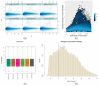
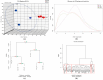
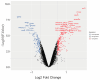
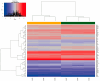
Patients with end-stage renal disease (ESRD) are at increased risk of cardiovascular disease (CVD), such as myocardial infarction (MI). Uremic toxins and endothelial dysfunction are central to this process. In this exploratory study, we used the Affymetrix GeneChip microarray to investigate the gene expression profile in uremic serum-induced human coronary arterial endothelial cells (HCAECs) from ESRD patients with and without MI (UWI and UWOI groups) as an approach to its underlying mechanism. We also explored which pathways are involved in this process. We found 100 differentially expressed genes (DEGs) among the conditions of interest by supervised principal component analysis and hierarchical cluster analysis. The expressions of four major DEGs were validated by quantitative RT-PCR. Pathway analysis and molecular network were used to analyze the interaction and expression patterns. Ten pathways were identified as the main enriched metabolic pathways according to the transcriptome profiling analysis, which were, among others, positive regulation of inflammatory response, positive regulation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) cascade, cardiac muscle cell development, highlighting positive regulation of mitogen-activated protein kinase (MAPK) activity (p = 0.00016). Up- and down-regulation of genes from HCAECs exposed to uremic serum could contribute to increased endothelial dysfunction and CVD in ESRD patients. Our study suggests that inflammation and the ERK-MAPK pathway are highly enriched in kidney disease patients with MI, suggesting their role in ESRD pathology. Further studies and approaches based on MAPK pathway interfering strategies are needed to confirm these data.
Keywords: chronic kidney disease; end-stage renal disease; endothelial cell dysfunction; gene expression profile; human coronary arterial endothelial cells; microarrays; myocardial infarction; uremia; uremic toxins.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Bikbov B., Purcell C., Levey A.S., Smith M., Abdoli A., Abebe M., Adebayo O.M., Afarideh M., Agarwal S.K., Ahmaidan E., et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2020;395:709–733. doi: 10.1016/S0140-6736(20)30045-3. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

