Decoding Immune Dynamics in Pregnant Women: Key Gene Expression Changes Following Influenza Vaccination
- PMID: 40332395
- PMCID: PMC12027590
- DOI: 10.3390/ijms26083765
Decoding Immune Dynamics in Pregnant Women: Key Gene Expression Changes Following Influenza Vaccination
Abstract
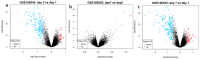
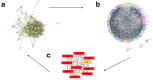
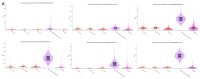
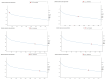
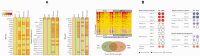
Pregnant women are at an increased risk of severe influenza complications, necessitating vaccination as a preventive measure. Despite World Health Organization (WHO) recommendations for influenza vaccination during pregnancy, vaccination rates remain suboptimal in many regions. This study aims to identify key differentially expressed genes (DEGs) and biological pathways modulated by influenza vaccination in pregnant women pre- and post-vaccination, contributing to improved vaccine strategies. Microarray data from gene expression omnibus GEO dataset GSE166545 was analyzed to identify DEGs in blood samples from pregnant women at three time points: pre-vaccination (Day 0) and post-vaccination (Days 0 and 1) (Days 1 and 7). DEGs were filtered using an adjusted p-value < 0.05 and |log2 fold change| ≥ 1. Protein/protein interaction (PPI) networks, hub gene identification, and pathway enrichment analyses were conducted using STRING, Cytoscape, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome databases. Hub gene validation was performed using the Human Protein Atlas (HPA) and GTEx Portal. The GSE166545 dataset analysis revealed 60 up-regulated and 12,854 down-regulated genes (Day 1 vs. 7), 55 up-regulated and 12,933 down-regulated genes (Day 0 vs. 1), and two up-regulated with no down-regulated genes (Day 0 vs. 7). Key pathways included interferon alpha/beta (IFN-γ\ β) signaling and toll-like receptor signaling (TLR). Hub genes such as GBP1, CXCL10, RSAD2, and IFI44 demonstrated robust up-regulation, correlating with enhanced immune responses. The initial observation of JCHAIN's notable up-regulation occurred on the seventh day following vaccination. Validation confirmed these genes' roles in antiviral defense mechanisms and vaccine responses. The findings reveal distinct immune response dynamics in pregnant women following influenza vaccination, highlighting potential biomarkers for vaccine efficacy. This study underscores the importance of tailored vaccine strategies to improve maternal and neonatal outcomes.
Keywords: hub genes; influenza; interferon pathway; pregnant women; transcriptomic analysis; vaccine.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Nakaya H.I., Hagan T., Duraisingham S.S., Lee E.K., Kwissa M., Rouphael N., Frasca D., Gersten M., Mehta A.K., Gaujoux R., et al. Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity. 2015;43:1186–1198. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- Project number (PNURSP2025R357)/Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R357), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
- project number NBU-FFR-2025-293-01./The authors extend their appreciation to the Deanship of Scientific Research at Northern Border University, Arar, KSA for funding this research work through the project number NBU-FFR-2025-293-01.
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

