MiR-22/GLUT1 Axis Induces Metabolic Reprogramming and Sorafenib Resistance in Hepatocellular Carcinoma
- PMID: 40332478
- PMCID: PMC12027541
- DOI: 10.3390/ijms26083808
MiR-22/GLUT1 Axis Induces Metabolic Reprogramming and Sorafenib Resistance in Hepatocellular Carcinoma
Abstract
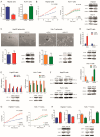
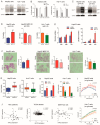
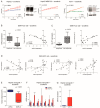
The approval of immunotherapy has revolutionized the management of hepatocellular carcinoma (HCC) patients. However, sorafenib remains a first-line therapeutic option for advanced patients and, in particular, for patients not eligible for immune checkpoint inhibitors, but its efficacy is limited by the onset of acquired resistance, highlighting the urgent need for predictive biomarkers. This study investigates the role of miR-22 in metabolic reprogramming and its potential as a biomarker in HCC. The analysis of miR-22 expression was performed in HCC patients and preclinical models by qPCR. Functional analyses in HCC cells evaluated GLUT1 as a direct miR-22 target. Cellular and metabolic assays evaluated the miR-22/GLUT1 axis's role in metabolic changes, tumor aggressiveness, and sorafenib response. Circulating miR-22 was analyzed in sorafenib-treated HCC patients and rats. MiR-22 was downregulated in HCCs and associated with aggressive tumor features. Functionally, miR-22 modulated the HIF1A pathway, enhanced survival in stressful conditions, promoted a glycolytic shift, and enhanced cancer cell plasticity and sorafenib resistance via GLUT1 targeting. In addition, high serum miR-22 levels were associated with sorafenib resistance in HCC patients and rats. GLUT1 inhibition sensitized low miR-22-expressing HCC cells to sorafenib in preclinical models. These findings suggest that circulating miR-22 deserves attention as a predictive biomarker of sorafenib response. GLUT1 inhibition may represent a therapeutic strategy to combine with sorafenib, particularly in patients exhibiting high circulating miR-22 levels.
Keywords: GLUT1; HCC; miR-22; sorafenib.
Conflict of interest statement
Indena S.p.A. has no commercial involvement with the study. The authors declare no conflicts of interest.
Figures



References
-
- Abou-Alfa G.K., Chan S.L., Kudo M., Lau G., Kelley R.K., Furuse J., Sukeepaisarnjaroen W., Kang Y.-K., Dao T.V., De Toni E.N., et al. Phase 3 Randomized, Open-Label, Multicenter Study of Tremelimumab (T) and Durvalumab (D) as First-Line Therapy in Patients (Pts) with Unresectable Hepatocellular Carcinoma (uHCC): HIMALAYA. J. Clin. Oncol. 2022;40:379. doi: 10.1200/JCO.2022.40.4_suppl.379. - DOI
-
- Reig M., Forner A., Rimola J., Ferrer-Fàbrega J., Burrel M., Garcia-Criado Á., Kelley R.K., Galle P.R., Mazzaferro V., Salem R., et al. BCLC Strategy for Prognosis Prediction and Treatment Recommendation: The 2022 Update. J. Hepatol. 2022;76:681–693. doi: 10.1016/j.jhep.2021.11.018. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- Development of 3D models, preclinical tools preclinical and circulating biomarkers for the optimization of personalized treatment approaches in hepatocellular carcinoma (SIMPLEMEDICINE)/University of Bologna
- IG-25187/Italian Association for Cancer Research
- PNRR - M4C2-I1.3 Project PE_00000019 "HEAL ITALIA" CUP of Institution: J33C22002920006./European Union
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

