scSDNE: A semi-supervised method for inferring cell-cell interactions based on graph embedding
- PMID: 40333631
- PMCID: PMC12072665
- DOI: 10.1371/journal.pcbi.1013027
scSDNE: A semi-supervised method for inferring cell-cell interactions based on graph embedding
Abstract
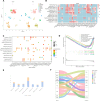
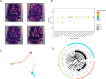
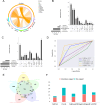
As a fundamental characteristic of multicellular organisms, cell-cell communication is achieved through ligand-receptor (L-R) interactions, enabling the exchange of information and revealing the diversity of biological processes and cellular functions. To gain a comprehensive understanding of these complex interaction mechanisms, we constructed a manually curated L-R interaction database and developed a semi-supervised graph embedding model called scSDNE for inferring cell-cell interactions mediated by L-R interactions. scSDNE model utilizes the power of deep learning to map genes from interacting cells into a shared latent space, allowing for a nuanced representation of their relationships. Leveraging the prior information provided by database, scSDNE can infer significant L-R pairs involved in intercellular communication. Experiments on real single-cell RNA sequencing (scRNA-seq) datasets demonstrate that our method detects interactions with a high degree of reliability compared with other methods. More importantly, the model integrates gene regulation information within cells to enhance the accuracy and biological interpretability of the inferences. Our method provides a more comprehensive view of cell-cell interactions, offering new insights into complex intercellular communication.
Copyright: © 2025 Jia et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

