Gene expression-based identification of prognostic markers in lung adenocarcinoma
- PMID: 40333815
- PMCID: PMC12057878
- DOI: 10.1371/journal.pone.0310232
Gene expression-based identification of prognostic markers in lung adenocarcinoma
Abstract
Introduction: Many studies have aimed at identifying additional prognostic tools to guide treatment choices and patient surveillance in lung cancer by assessing the expression of individual proteins through immunohistochemistry (IHC) or, more recently, through gene expression-based signatures. As a proof-of-concept, we used a multi-cohort, gene expression-based discovery and validation strategy to identify genes with prognostic potential in lung adenocarcinoma. The clinical applicability of this strategy was further assessed by evaluating a selection of the markers by IHC.
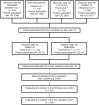
Materials and methods: Publicly available gene expression data sets from six microarray-based studies were divided into four discovery and two validation data sets. First, genes associated with overall survival (OS) in all four discovery data sets were identified. The prognostic potential of each identified gene was then assessed in the two validation data sets, and genes associated with OS in both data sets were considered as potential prognostic markers. Finally, IHC for selected potential prognostic markers was performed in two independent and clinically well-characterized lung cancer cohorts.
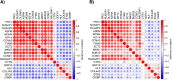
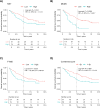
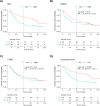
Results and conclusions: The gene expression-based strategy identified 19 genes with correlation to OS in all six data sets. Out of these genes, we selected Ki67, MCM4 and TYMS for further assessment with IHC. Although an independent prognostic ability of the selected markers could not be confirmed by IHC, this proof-of-concept study demonstrates that by employing a gene expression-based discovery and validation strategy, potential prognostic markers can be identified and further assessed by a technique universally applicable in the clinical practice. The concept of studying potential prognostic markers through gene expression-based strategies, with a subsequent evaluation of the clinical utility, warrants further exploration.
Copyright: © 2025 Salomonsson et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Felip E, Altorki N, Zhou C, Csőszi T, Vynnychenko I, Goloborodko O, et al.. Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB-IIIA non-small-cell lung cancer (IMpower010): a randomised, multicentre, open-label, phase 3 trial. Lancet. 2021;398(10308):1344–57. doi: 10.1016/S0140-6736(21)02098-5 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

