C16orf74 is a novel prognostic biomarker and associates with immune infiltration in head and neck squamous cell carcinoma
- PMID: 40333816
- PMCID: PMC12057912
- DOI: 10.1371/journal.pone.0322701
C16orf74 is a novel prognostic biomarker and associates with immune infiltration in head and neck squamous cell carcinoma
Abstract
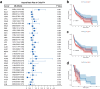
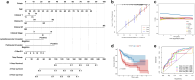
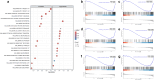
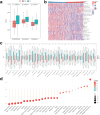
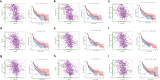
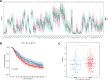
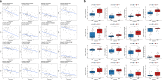
Head and neck squamous cell carcinoma (HNSC) is a prevalent and aggressive malignancy with poor prognosis, underscoring the need for novel biomarkers and therapeutic strategies. This study investigates the role of C16orf74 as a potential diagnostic and prognostic biomarker in HNSC. Bioinformatics analyses revealed that C16orf74 is significantly overexpressed in HNSC and is associated with advanced disease stages, therapy resistance, and shorter overall and progression-free survival. A prognostic nomogram integrating C16orf74 expression with clinicopathological features demonstrated robust predictive performance. Functional enrichment and immune infiltration analyses suggest that high C16orf74 expression might contribute to an immunosuppressive tumor microenvironment by reducing key immune cell populations, such as B cells, T cells, and natural killer cells, which are critical for anti-tumor immunity. Moreover, C16orf74 expression was inversely associated with immune checkpoint expression and immunotherapy response, highlighting its potential as a predictive biomarker for immune checkpoint blockade (ICB) efficacy. Drug sensitivity analyses identified potential therapeutic agents, including arsenic trioxide, carmustine, vincristine, quercetin, and carboplatin for patients with high C16orf74 expression. These findings highlight the potential of C16orf74 as a biomarker and therapeutic target to improve HNSC management.
Copyright: © 2025 Yao et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

