Niemann Pick C1 mistargeting disrupts lysosomal cholesterol homeostasis contributing to neurodegeneration in a Batten disease model
- PMID: 40333988
- PMCID: PMC12057685
- DOI: 10.1126/sciadv.adr5703
Niemann Pick C1 mistargeting disrupts lysosomal cholesterol homeostasis contributing to neurodegeneration in a Batten disease model
Abstract
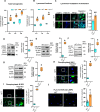
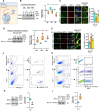
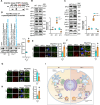
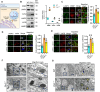
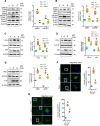
Neurodegeneration is a devastating manifestation in most lysosomal storage disorders (LSDs). Loss-of-function mutations in CLN1, encoding palmitoyl-protein thioesterase-1 (PPT1), cause CLN1 disease, a devastating neurodegenerative LSD that has no curative treatment. Numerous proteins in the brain require dynamic S-palmitoylation (palmitoylation-depalmitoylation) for trafficking to their destination. Although PPT1 depalmitoylates S-palmitoylated proteins and its deficiency causes CLN1 disease, the underlying pathogenic mechanism has remained elusive. We report that Niemann-Pick C1 (NPC1), a polytopic membrane protein mediating lysosomal cholesterol egress, requires dynamic S-palmitoylation for trafficking to the lysosome. In Cln1-/- mice, Ppt1 deficiency misroutes NPC1-dysregulating lysosomal cholesterol homeostasis. Along with this defect, increased oxysterol-binding protein (OSBP) promotes cholesterol-mediated activation of mechanistic target of rapamycin C1 (mTORC1), which inhibits autophagy contributing to neurodegeneration. Pharmacological inhibition of OSBP suppresses mTORC1 activation, rescues autophagy, and ameliorates neuropathology in Cln1-/- mice. Our findings reveal a previously unrecognized role of CLN1/PPT1 in lysosomal cholesterol homeostasis and suggest that suppression of mTORC1 activation may be beneficial for CLN1 disease.
Figures
References
-
- de Duve C., Wattiaux R., Functions of lysosomes. Annu. Rev. Physiol. 28, 435–492 (1966). - PubMed
-
- Saftig P., Klumperman J., Lysosome biogenesis and lysosomal membrane proteins: Trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635 (2009). - PubMed
-
- Marques A. R. A., Saftig P., Lysosomal storage disorders - challenges, concepts and avenues for therapy: Beyond rare diseases. J. Cell Sci. 132, jcs221739 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

