Identification of Distinct Biological Groups of Patients With Cryptogenic NORSE via Inflammatory Profiling
- PMID: 40334176
- PMCID: PMC12063244
- DOI: 10.1212/NXI.0000000000200403
Identification of Distinct Biological Groups of Patients With Cryptogenic NORSE via Inflammatory Profiling
Abstract
Background and objectives: Emerging evidence suggests that immune dysregulation plays a pivotal role in triggering cryptogenic new-onset refractory status epilepticus (c-NORSE), prompting a consensus on early initiation of immunotherapy. However, despite similar timing of administration, responses to immunotherapies have been varied and unpredictable, suggesting the presence of heterogeneous underlying mechanisms. The aim of this study was to identify distinct inflammatory response subtypes in patients with c-NORSE by analyzing their cytokine profiles. Insights into underlying mechanisms were sought to understand the pathophysiology and guide personalized therapies to improve patient outcomes.
Methods: Sixty-two patients with c-NORSE were included. A comprehensive panel of 96 cytokines was analyzed in serum samples. Patients were clustered based on their cytokine profiles using the Louvain algorithm, an unsupervised graph-based clustering method. The identified clusters of patients were compared regarding cytokine levels and clinical features. Protein pathway analysis was used to explore the biological relevance of the inflammatory markers within each cluster. Patients with c-NORSE were compared with control patients (n = 18) and patients with other forms of refractory SE (n = 45).
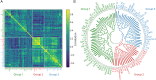
Results: Compared with controls, patients with c-NORSE exhibited significant differences in 33 cytokines. Pathway analysis revealed dysregulations in chemotaxis and neutrophil recruitment and migration, highlighting the importance of innate immunity in patients with c-NORSE. Within the c-NORSE cohort, 3 clusters of patients emerged: cluster A, lacking specific inflammatory markers; cluster B, with a much stronger innate-immunity cytokine-driven inflammatory response compared with clusters A and C; and cluster C, defined by dysregulated autoimmune processes. Notably, patients in cluster B showed a statistically significant elevation of innate immune-related proinflammatory cytokines associated with leukocyte recruitment and degranulation. By contrast, those in cluster C showed activation of Janus kinase signal transducer and activator of transcription (JAK-STAT) pathways, suggesting autoimmune mechanisms. Patients in clusters B and C demonstrated varied responses to immunotherapies, with cluster C patients showing favorable outcomes after multiple immunotherapies.
Discussion: The identification of distinct inflammatory subgroups in c-NORSE suggests that variations in the underlying immune mechanisms contribute to differential treatment responses. These findings underscore the importance of personalized therapeutic strategies, potentially targeting specific inflammatory pathways, to optimize clinical outcomes in this challenging condition.
Conflict of interest statement
L.J. Hirsch received support to Yale University for investigator-initiated studies from The Daniel Raymond Wong Neurology Research Fund and the NORSE/FIRES Research Fund at Yale. A. Hanin received postdoctoral grants from the Paratonnerre Association, the Swebilius Foundation, the Servier Institute, and the Philippe Foundation for NORSE-related research. A. Vezzani, K. Eschbach, N. Gaspard, T.E. Gofton, H.A. Haider, C.L. Howe, Y.-C. Lai,O. Taraschenko, L.J. Hirsch, and A. Hanin are members of the Medical and Scientific Advisory Board of the NORSE Institute. K. Eschbach received funding from the friends and family of Finn Mussetter for NORSE and FIRES research. H.A. Haider received in 2020–2022 philanthropic funding from the Neal Nichols Foundation for Status Epilepticus/NORSE Research. V. Navarro received a CURE Epilepsy grant in 2024 for NORSE-related research. The other authors report no competing interests. Go to
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
