Do we need a standardized 16S rRNA gene amplicon sequencing analysis protocol for poultry microbiota research?
- PMID: 40334389
- PMCID: PMC12207820
- DOI: 10.1016/j.psj.2025.105242
Do we need a standardized 16S rRNA gene amplicon sequencing analysis protocol for poultry microbiota research?
Abstract
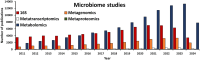
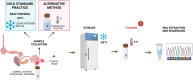
Bacteria are the major component of poultry gastrointestinal tract (GIT) microbiota and play an important role in host health, nutrition, physiology regulation, intestinal development, and growth. Bacterial community profiling based on the 16S ribosomal RNA (rRNA) gene amplicon sequencing approach has become the most popular method to determine the taxonomic composition and diversity of the poultry microbiota. The 16S rRNA gene profiling involves numerous steps, including sample collection and storage, DNA isolation, 16S rRNA gene primer selection, Polymerase Chain Reaction (PCR), library preparation, sequencing, raw sequencing reads processing, taxonomic classification, α- and β-diversity calculations, and statistical analysis. However, there is currently no standardized protocol for 16S rRNA gene analysis profiling and data deposition for poultry microbiota studies. Variations in DNA storage and isolation, primer design, and library preparation are known to introduce biases, affecting community structure and microbial population analysis leading to over- or under-representation of individual bacteria within communities. Additionally, different sequencing platforms, bioinformatics pipeline, and taxonomic database selection can affect classification and determination of the microbial taxa. Moreover, detailed experimental design and DNA processing and sequencing methods are often inadequately reported in poultry 16S rRNA gene sequencing studies. Consequently, poultry microbiota results are often difficult to reproduce and compare across studies. This manuscript reviews current practices in profiling poultry microbiota using 16S rRNA gene amplicon sequencing and proposes the development of guidelines for protocol for 16S rRNA gene sequencing that spans from sample collection through data deposition to achieve more reliable data comparisons across studies and allow for comparisons and/or interpretations of poultry studies conducted worldwide.
Keywords: 16S rRNA gene sequencing; Microbiota; Next-generation sequencing; Poultry; Standardized protocol.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Disclosures The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Aitchison J. Chapman & Hall Ltd.; London (UK): 1986. The Statistical Analysis of Compositional Data Monographs on Statistics and Applied Probability.
-
- Allali I., Arnold J.W., Roach J., Cadenas M.B., Butz N., Hassan H.M., Koci M., Ballou A., Mendoza M., Ali R., Azcarate-Peril M.A. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC. Microbiol. 2017;17:194. doi: 10.1186/s12866-017-1101-8. - DOI - PMC - PubMed
-
- Allali I., Arnold J.W., Roach J., Cadenas M.B., Butz N., Hassan H.M., Koci M., Ballou A., Mendoza M., Ali R., Azcarate-Peril M.A. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC. Microbiol. 2017;17:194. doi: 10.1186/s12866-017-1101-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

