Discovery of Proteoforms Associated With Alzheimer's Disease Through Quantitative Top-Down Proteomics
- PMID: 40334744
- PMCID: PMC12173667
- DOI: 10.1016/j.mcpro.2025.100983
Discovery of Proteoforms Associated With Alzheimer's Disease Through Quantitative Top-Down Proteomics
Abstract
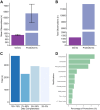
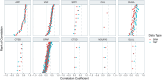
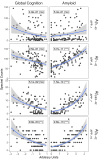
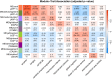
The complex nature of Alzheimer's disease (AD) and its heterogenous clinical presentation has prompted numerous large-scale -omic analyses aimed at providing a global understanding of the pathophysiological processes involved. AD involves isoforms, proteolytic products, and posttranslationally modified proteins such as amyloid beta (Aβ) and microtubule-associated protein tau. Top-down proteomics directly measures these species and thus, offers a comprehensive view of pathologically relevant proteoforms that are difficult to analyze using traditional proteomic techniques. Here, we broadly explored associations between proteoforms and clinicopathological traits of AD by deploying a quantitative top-down proteomics approach across frontal cortex of 103 subjects selected from the ROS and MAP cohorts. The approach identified 1213 proteins and 11,782 proteoforms, of which 154 proteoforms had at least one significant association with a clinicopathological phenotype. One important finding included identifying Aβ C-terminal truncation state as the key property for differential association between amyloid plaques and cerebral amyloid angiopathy. Furthermore, various N-terminally truncated forms of Aβ had noticeably stronger association with amyloid plaques and global cognitive function. Additionally, we discovered six VGF neuropeptides that were positively associated with cognitive function independent of pathological burden. The database of brain cortex proteoforms provides a valuable context for functional characterization of the proteins involved in AD and other late-onset brain pathologies.
Keywords: Alzheimer's disease; amyloid beta; top-down proteomics.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interests The authors declare that they have no conflicts of interests with the contents of this article.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

