Molecular evolution of a reproductive barrier in maize and related species
- PMID: 40335053
- PMCID: PMC12239200
- DOI: 10.1093/genetics/iyaf085
Molecular evolution of a reproductive barrier in maize and related species
Abstract
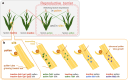
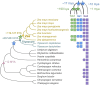
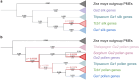
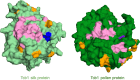
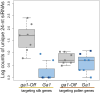
Three cross-incompatibility loci each control a distinct reproductive barrier in both domesticated maize (Zea mays ssp. mays) and its wild teosinte relatives. These 3 loci, Teosinte crossing barrier1 (Tcb1), Gametophytic factor1 (Ga1), and Ga2, each play a key role in preventing hybridization between incompatible populations and are proposed to maintain the barrier between domesticated and wild subspecies. Each locus encodes both a silk-active and a matching pollen-active pectin methylesterase (PMEs). To investigate the diversity and molecular evolution of these gametophytic factor loci, we identified existing and improved models of the responsible genes in a new genome assembly of maize line P8860 that contains active versions of all 3 loci. We then examined 52 assembled genomes from 17 species to classify haplotype diversity and identify sites under diversifying selection during the evolution of these genes. We show that Ga2, the oldest of these 3 loci, was duplicated to form Ga1 at least 12 million years ago. Tcb1, the youngest locus, arose as a duplicate of Ga1 before or around the time of diversification of the Zea genus. We find evidence of positive selection during evolution of the functional genes at an active site in the pollen-expressed PME and predicted surface sites in both the silk- and pollen-expressed PMEs. The most common allele at the Ga1 locus is a conserved ga1 allele (ga1-Off), which is specific haplotype containing 3 full-length PME gene copies, all of which are noncoding due to conserved stop codons and are between 610 thousand and 1.5 million years old. We show that the ga1-Off allele is associated with and likely generates 24-nt siRNAs in developing pollen-producing tissue, and these siRNAs map to functional Ga1 alleles. In previously published crosses, the ga1-Off allele was associated with reduced function of the typically dominant functional alleles for the Ga1 and Tcb1 barriers. Taken together, this seems to be an example of an allele at a reproductive barrier locus being associated with an as yet undetermined mechanism capable of silencing the reproductive barrier.
Keywords: gametophytic factors; genome assembly; maize; molecular evolution; pollen; reproductive barrier; siRNA; silencing; silk; transmission ratio distortion.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The authors declare no conflict of interest.
Figures


References
-
- Ashman RB. 1975. Modification of cross-sterility in maize. J Hered. 66(1):5–9. doi: 10.1093/oxfordjournals.jhered.a108577. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

