Spatial transcriptional landscape of human heart failure
- PMID: 40335066
- PMCID: PMC12349961
- DOI: 10.1093/eurheartj/ehaf272
Spatial transcriptional landscape of human heart failure
Abstract
Background and aims: Heart failure (HF) remains a significant clinical challenge due to its diverse aetiologies and complex pathophysiology. The molecular alterations specific to distinct cell types and histological patterns during HF progression are still poorly characterized. This study aimed to explore cell-type- and histology-specific gene expression profiles in cardiomyopathies.
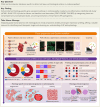
Methods: Ninety tissue cores from 44 participants, encompassing various forms of cardiomyopathy and control samples with diverse histological features, were analysed using the GeoMx Whole Human Transcriptome Atlas. Data on cell types, clinical information, and histological features were integrated to examine gene expression profiles in cardiomyopathy.
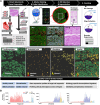
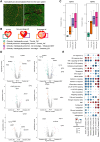
Results: The study characterized the cellular composition of ventricular myocardium and validated the GeoMx platform's efficiency in compartmentalizing specific cell types, demonstrating high accuracy for cardiomyocytes but limitations for endothelial cells and fibroblasts. Differentially expressed genes, including UCHL1 from cardiomyocytes, were associated with degeneration, while CCL14, ACKR1, and PLVAP from endothelial cells were linked to fibrosis. Multiplex immunohistochemistry and integrative analysis of prior sc/snRNA-seq data identified a PLVAP, ACKR1, and CCL14-positive pro-inflammatory endothelial cell subtype linked to fibrosis in HF. Downregulation of ribosomal proteins in cardiomyocytes was associated with myocyte disarray in hypertrophic cardiomyopathy. Additionally, pronounced inflammatory responses were observed in end-stage HF. Combined histological and clinical analysis identified CRIP3, PFKFB2, and TAX1BP3 as novel contributors to HF pathogenesis.
Conclusions: These findings highlight the critical role of cell-enriched and histology-specific transcriptome mapping in understanding the complex pathophysiological landscape of failing hearts, offering molecular insights and potential therapeutic targets for future interventions.
Keywords: Cardiomyocytes; Cardiomyopathy; Degeneration; Endothelial cells; Fibrosis; Heart failure; Inflammation; Myocyte disarray; Spatial transcriptomics.
© The Author(s) 2025. Published by Oxford University Press on behalf of the European Society of Cardiology.
Figures




References
-
- Heidenreich PA, Bozkurt B, Aguilar D, Allen LA, Byun JJ, Colvin MM, et al. 2022 AHA/ACC/HFSA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2022;145:E895–E1032. 10.1161/CIR.0000000000001063 - DOI - PubMed
-
- Burnett H, Earley A, Voors AA, Senni M, McMurray JJV, Deschaseaux C, et al. Thirty years of evidence on the efficacy of drug treatments for chronic heart failure with reduced ejection fraction: a network meta-analysis. Circ Heart Fail 2017;10:e003529. 10.1161/CIRCHEARTFAILURE.116.003529/-/DC1 - DOI - PMC - PubMed
-
- Lippi G, Sanchis-Gomar F. Global epidemiology and future trends of heart failure. AME Med J 2020;5:15. 10.21037/AMJ.2020.03.03 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

