Sirt6 deficiency promotes senescence and age-associated intervertebral disc degeneration in mice
- PMID: 40335469
- PMCID: PMC12059161
- DOI: 10.1038/s41413-025-00422-3
Sirt6 deficiency promotes senescence and age-associated intervertebral disc degeneration in mice
Abstract
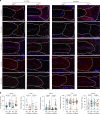
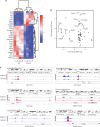
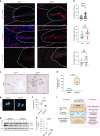
Intervertebral disc degeneration is a major risk factor contributing to chronic low back and neck pain. While the etiological factors for disc degeneration vary, age is still one of the most important risk factors. Recent studies have shown the promising role of SIRT6 in mammalian aging and skeletal tissue health, however its role in the intervertebral disc health remains unexplored. We investigated the contribution of SIRT6 to disc health by studying the age-dependent spinal phenotype of mice with conditional deletion of Sirt6 in the disc (AcanCreERT2; Sirt6fl/fl). Histological studies showed a degenerative phenotype in knockout mice compared to Sirt6fl/fl control mice at 12 months, which became pronounced at 24 months. RNA-Seq analysis of NP and AF tissues, in vitro quantitative histone analysis, and RNA-seq with ATAC-seq multiomic studies revealed that SIRT6-loss resulted in changes in acetylation and methylation status of specific Histone 3 lysine residues and affected DNA accessibility and transcriptomic landscape. A decrease in autophagy and an increase in DNA damage were also noted in Sirt6-deficient cells. Further mechanistic insights revealed that loss of SIRT6 increased senescence and SASP burden in the disc characterized by increased p21, p19, γH2AX, IL-6, IL-1β, and TGF-β abundance. Taken together, our study highlights the contribution of SIRT6 in modulating DNA damage, autophagy, and cell senescence and its importance in maintaining disc health during aging, thereby underscoring it as a potential therapeutic target to treat intervertebral disc degeneration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics: All animal experiments were performed under IACUC protocols approved by the University of North Carolina at Chapel Hill and Thomas Jefferson University.
Figures




Update of
-
SIRT6 loss causes intervertebral disc degeneration in mice by promoting senescence and SASP status.bioRxiv [Preprint]. 2024 Sep 9:2024.09.09.612072. doi: 10.1101/2024.09.09.612072. bioRxiv. 2024. Update in: Bone Res. 2025 May 8;13(1):50. doi: 10.1038/s41413-025-00422-3. PMID: 39314282 Free PMC article. Updated. Preprint.
References
-
- Farsetti, A., Illi, B. & Gaetano, C. How epigenetics impacts on human diseases. Eur. J. Intern. Med.114, 15–22 (2023). - PubMed
MeSH terms
Substances
Grants and funding
- R01 AG078609/AG/NIA NIH HHS/United States
- R01 AG044034/AG/NIA NIH HHS/United States
- R01AG044034/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01AG073349/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01 AG073349/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

