Activation of lysosomal iron triggers ferroptosis in cancer
- PMID: 40335696
- PMCID: PMC12158755
- DOI: 10.1038/s41586-025-08974-4
Activation of lysosomal iron triggers ferroptosis in cancer
Abstract
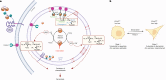
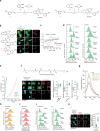
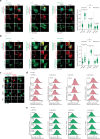
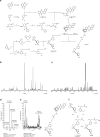
Iron catalyses the oxidation of lipids in biological membranes and promotes a form of cell death called ferroptosis1. Defining where this chemistry occurs in the cell can inform the design of drugs capable of inducing or inhibiting ferroptosis in various disease-relevant settings. Genetic approaches have revealed suppressors of ferroptosis2-4; by contrast, small molecules can provide spatiotemporal control of the chemistry at work5. Here we show that the ferroptosis inhibitor liproxstatin-1 exerts cytoprotective effects by inactivating iron in lysosomes. We also show that the ferroptosis inducer RSL3 initiates membrane lipid oxidation in lysosomes. We designed a small-molecule activator of lysosomal iron-fentomycin-1-to induce the oxidative degradation of phospholipids and ultimately ferroptosis. Fentomycin-1 is able to kill iron-rich CD44high primary sarcoma and pancreatic ductal adenocarcinoma cells, which can promote metastasis and fuel drug tolerance. In such cells, iron regulates cell adaptation6,7 while conferring vulnerability to ferroptosis8,9. Sarcoma cells exposed to sublethal doses of fentomycin-1 acquire a ferroptosis-resistant cell state characterized by the downregulation of mesenchymal markers and the activation of a membrane-damage response. This phospholipid degrader can eradicate drug-tolerant persister cancer cells in vitro and reduces intranodal tumour growth in a mouse model of breast cancer metastasis. Together, these results show that control of iron reactivity confers therapeutic benefits, establish lysosomal iron as a druggable target and highlight the value of targeting cell states10.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Institut Curie filed a patent application (PCT/EP2025/051999) related to lipid degraders to induce ferroptosis in cancer. M.C. and B.P. hold patents for some of the compounds described herein and are co-founders of ROSCUE Therapeutics. B.R.S. is an inventor on patents and patent applications involving ferroptosis; co-founded and serves as a consultant to ProJenX and Exarta Therapeutics; holds equity in Sonata Therapeutics; and serves as a consultant to Weatherwax Biotechnologies and Akin Gump Strauss Hauer & Feld. All the other authors declare no competing interests.
Figures









Update of
-
Activation of lysosomal iron triggers ferroptosis in cancer.Res Sq [Preprint]. 2024 Apr 8:rs.3.rs-4165774. doi: 10.21203/rs.3.rs-4165774/v1. Res Sq. 2024. Update in: Nature. 2025 Jun;642(8067):492-500. doi: 10.1038/s41586-025-08974-4. PMID: 38659936 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

