Oncogene aberrations drive medulloblastoma progression, not initiation
- PMID: 40335697
- PMCID: PMC12222029
- DOI: 10.1038/s41586-025-08973-5
Oncogene aberrations drive medulloblastoma progression, not initiation
Abstract
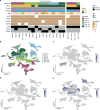
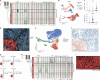
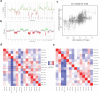
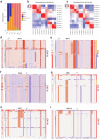
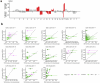
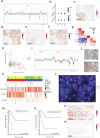
Despite recent advances in understanding disease biology, treatment of group 3/4 medulloblastoma remains a therapeutic challenge in paediatric neuro-oncology1. Bulk-omics approaches have identified considerable intertumoural heterogeneity in group 3/4 medulloblastoma, including the presence of clear single-gene oncogenic drivers in only a subset of cases, whereas in most cases, large-scale copy number aberrations prevail2,3. However, intratumoural heterogeneity, the role of oncogene aberrations, and broad copy number variation in tumour evolution and treatment resistance remain poorly understood. To dissect this interplay, we used single-cell technologies (single-nucleus RNA sequencing (snRNA-seq), single-nucleus assay for transposase-accessible chromatin with high-throughput sequencing (snATAC-seq) and spatial transcriptomics) on a cohort of group 3/4 medulloblastoma with known alterations in the oncogenes MYC, MYCN and PRDM6. We show that large-scale chromosomal aberrations are early tumour-initiating events, whereas the single-gene oncogenic events arise late and are typically subclonal, but MYC can become clonal upon disease progression to drive further tumour development and therapy resistance. Spatial transcriptomics shows that the subclones are mostly interspersed across tumour tissue, but clear segregation is also present. Using a population genetics model, we estimate medulloblastoma initiation in the cerebellar unipolar brush cell lineage starting from the first gestational trimester. Our findings demonstrate how single-cell technologies can be applied for early detection and diagnosis of this fatal disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: C.M.v.T. participates on advisory boards for Alexion, Bayer and Novartis. The other authors declare no competing interests.
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

