PLA2G15 is a BMP hydrolase and its targeting ameliorates lysosomal disease
- PMID: 40335701
- PMCID: PMC12158761
- DOI: 10.1038/s41586-025-08942-y
PLA2G15 is a BMP hydrolase and its targeting ameliorates lysosomal disease
Abstract
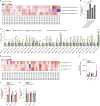
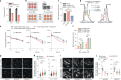
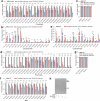
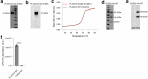
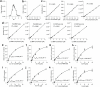
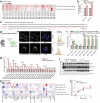
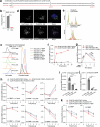
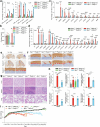
Lysosomes catabolize lipids and other biological molecules, maintaining cellular and organismal homeostasis. Bis(monoacylglycero)phosphate (BMP), a major lipid constituent of intralysosomal vesicles, stimulates lipid-degrading enzymes and is altered in various human conditions, including neurodegenerative diseases1,2. Although lysosomal BMP synthase was recently discovered3, the enzymes mediating BMP turnover remain elusive. Here we show that lysosomal phospholipase PLA2G15 is a physiological BMP hydrolase. We further demonstrate that the resistance of BMP to lysosomal hydrolysis arises from its unique sn2, sn2' esterification position and stereochemistry, as neither feature alone confers resistance. Purified PLA2G15 catabolizes most BMP species derived from cell and tissue lysosomes. Furthermore, PLA2G15 efficiently hydrolyses synthesized BMP stereoisomers with primary esters, challenging the long-held thought that BMP stereochemistry alone ensures resistance to acid phospholipases. Conversely, BMP with secondary esters and S,S stereoconfiguration is stable in vitro and requires acyl migration for hydrolysis in lysosomes. Consistent with our biochemical data, PLA2G15-deficient cells and tissues accumulate several BMP species, a phenotype reversible by supplementing wild-type PLA2G15 but not its inactive mutant. Targeting PLA2G15 reduces the cholesterol accumulation in fibroblasts of patients with Niemann-Pick disease type C1 and significantly ameliorates disease pathologies in Niemann-Pick disease type C1-deficient mice, leading to an extended lifespan. Our findings established the rules governing BMP stability in lysosomes and identified PLA2G15 as a lysosomal BMP hydrolase and a potential target for therapeutic intervention in neurodegenerative diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.A.-R. is a scientific advisory board member of Lycia Therapeutics and senior advisor of Scenic Biotech. M.R., V.A.B., S.M.B.N., T.R.B., A.P.H.d.J. and G.H. are employees or advisors of Scenic Biotech. R.d.M. is an employee of AnaPath Services. The other authors declare no competing interests.
Figures







Update of
-
PLA2G15 is a Lysosomal BMP Hydrolase and its Targeting Ameliorates Lysosomal Disease.bioRxiv [Preprint]. 2025 Feb 19:2024.06.07.597919. doi: 10.1101/2024.06.07.597919. bioRxiv. 2025. Update in: Nature. 2025 Jun;642(8067):474-483. doi: 10.1038/s41586-025-08942-y. PMID: 38895439 Free PMC article. Updated. Preprint.
References
-
- Medoh, U. N. & Abu-Remaileh, M. The bis(monoacylglycero)-phosphate hypothesis: from lysosomal function to therapeutic avenues. Annu. Rev. Biochem. 10.1146/annurev-biochem-092823-113814 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

