Genome-wide association meta-analysis of age at onset of walking in over 70,000 infants of European ancestry
- PMID: 40335706
- PMCID: PMC12283345
- DOI: 10.1038/s41562-025-02145-1
Genome-wide association meta-analysis of age at onset of walking in over 70,000 infants of European ancestry
Abstract
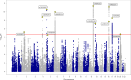
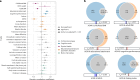
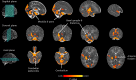
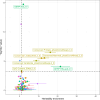
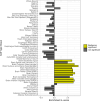
Age at onset of walking is an important early childhood milestone which is used clinically and in public health screening. In this genome-wide association study meta-analysis of age at onset of walking (N = 70,560 European-ancestry infants), we identified 11 independent genome-wide significant loci. SNP-based heritability was 24.13% (95% confidence intervals = 21.86-26.40) with ~11,900 variants accounting for about 90% of it, suggesting high polygenicity. One of these loci, in gene RBL2, co-localized with an expression quantitative trait locus (eQTL) in the brain. Age at onset of walking (in months) was negatively genetically correlated with ADHD and body-mass index, and positively genetically correlated with brain gyrification in both infant and adult brains. The polygenic score showed out-of-sample prediction of 3-5.6%, confirmed as largely due to direct effects in sib-pair analyses, and was separately associated with volume of neonatal brain structures involved in motor control. This study offers biological insights into a key behavioural marker of neurodevelopment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: O.A.A. is a consultant to cortechs.ai and Precision Health, and receives speaker honoraria from Janssen, Lundbeck, Sunovion, Lilly, and Otsuka. S.J.S. receives research funding from BioMarin Pharmaceutical. The other authors declare no competing interests.
Figures


References
-
- Langendonk, J. M. et al. Assessment of motor milestones in twins. Twin Res. Hum. Genet.10, 835–839 (2007). - PubMed
-
- Help your child grow and thrive. Centers for Disease Control and Preventionhttps://www.cdc.gov/ncbddd/actearly/index.html (2025).
-
- WHO Multicentre Growth Reference Study Group WHO Motor Development Study: windows of achievement for six gross motor development milestones. Acta Paediatr. 95, 86–95 (2006). - PubMed
-
- Bowler, A., Arichi, T., Austerberry, C., Fearon, P. & Ronald, A. A systematic review and meta-analysis of the associations between motor milestone timing and motor development in neurodevelopmental conditions. Neurosci. Biobehav. Rev.167, 105825 (2024). - PubMed
Publication types
MeSH terms
Grants and funding
- RC2 MH089995/MH/NIMH NIH HHS/United States
- R01 MH129751/MH/NIMH NIH HHS/United States
- MR/S037055/1/RCUK | Medical Research Council (MRC)
- MR/V036874/1/RCUK | Medical Research Council (MRC)
- 324252/Norges Forskningsråd (Research Council of Norway)
- R01 MH058799/MH/NIMH NIH HHS/United States
- EP-C-15-001/EPA/EPA/United States
- U24 MH068457/MH/NIMH NIH HHS/United States
- 164218/NordForsk
- RC2 MH089951/MH/NIMH NIH HHS/United States
- MR/T003057/RCUK | Medical Research Council (MRC)
- 223273/Norges Forskningsråd (Research Council of Norway)
- R01 MH081802/MH/NIMH NIH HHS/United States
- ES/P000592/1/RCUK | Economic and Social Research Council (ESRC)
- 724306/Simons Foundation
- U01 MH122681/MH/NIMH NIH HHS/United States
- 324499/Norges Forskningsråd (Research Council of Norway)
- 274611/Norges Forskningsråd (Research Council of Norway)
- 336085/Norges Forskningsråd (Research Council of Norway)
- 101057385/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- MR/N026063/1/RCUK | Medical Research Council (MRC)
- R01MH129751/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R01 DK092127/DK/NIDDK NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U01MH122681/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- MC_UU_00019/1/RCUK | Medical Research Council (MRC)
- 214322\Z\18\Z/Wellcome Trust (Wellcome)
- 964874/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 101073237/EC | EU Framework Programme for Research and Innovation H2020 | H2020 European Institute of Innovation and Technology (H2020 The European Institute of Innovation and Technology)
LinkOut - more resources
Full Text Sources

