Genomic surveillance reveals different transmission patterns between third-generation cephalosporin and carbapenem resistance in Klebsiella pneumoniae in the Comunidad Valenciana (Spain), 2018-2020
- PMID: 40336117
- PMCID: PMC12060429
- DOI: 10.1186/s13756-025-01553-2
Genomic surveillance reveals different transmission patterns between third-generation cephalosporin and carbapenem resistance in Klebsiella pneumoniae in the Comunidad Valenciana (Spain), 2018-2020
Abstract
Background: The emergence and spread of third-generation cephalosporins (3GC) and carbapenem-resistant Klebsiella pneumoniae pose a global critical challenge. Understanding the transmission dynamics within and between hospital environments is crucial to develop effective control strategies.
Methods: From 2017 to 2019, we conducted a genomic surveillance program in eight hospitals of the Comunitat Valenciana, Spain, collecting and sequencing 1,768 3GC- and carbapenem-resistant isolates. We quantified the overall transmission using core genomes and assessed the contribution of national and global isolates to the spread of AMR in the region by including 11,967 database genomes in the analysis.
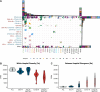
Results: The local collection was highly diverse, involving 188 lineages, including global high-risk clones such as ST307 and ST11, and 3GC and carbapenem resistance determinants. Half of the isolates were involved in transmission, with 70.5% occurring within hospitals.
Conclusions: Different transmission patterns characterized the spread of 3GC- and carbapenem resistance in the region. While inter-hospital transmission played a significant role in the spread of 3GC-resistance, this was only sporadic for carbapenem resistance. Moreover, the factors behind inter-hospital spread for each type of resistance differed: while 3GC-resistance likely disseminated between hospitals through intermediate steps, carbapenem resistance was driven by more direct transmission routes. The burden of national and global cases on the ongoing regional AMR dissemination was low. Moreover, we revealed the rapid expansion in the region and globally of lineage ST307 carrying the blaCTX-M-15 gene, a main driver of local transmissions, providing a deeper understanding of the successful spread of this high-risk clone.
Keywords: Klebsiella pneumoniae; Carbapenem resistance; Genomic surveillance.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The results reported make no use of individual information and were obtained under a routine surveillance project which the DGSP-FISABIO ethics committee declared to be exempt from approval. Competing interests: The authors declare no competing interests.
Figures





References
-
- World Health Organization. WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. 2024. https://www.who.int/publications/i/item/9789240093461. Accessed 13 Mar 2024.
-
- Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18:344–59. - PubMed
-
- Chong Y, Shimoda S, Yakushiji H, et al. Community spread of extended-spectrum β-lactamase-producing Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis: a long-term study in Japan. J Med Microbiol. 2013;62:1038–43. - PubMed
MeSH terms
Substances
Grants and funding
- PID2021-127010OB-I00/Ministerio de Ciencia e Innovación
- PID2021-127010OB-I00/Ministerio de Ciencia e Innovación
- PID2021-127010OB-I00/Ministerio de Ciencia e Innovación
- PID2021-127010OB-I00/Ministerio de Ciencia e Innovación
- CIPROM-2021-053/Conselleria de Cultura, Educación y Ciencia, Generalitat Valenciana
LinkOut - more resources
Full Text Sources
Medical

