Temporal transcriptomic changes in the THY-Tau22 mouse model of tauopathy display cell type- and sex-specific differences
- PMID: 40336141
- PMCID: PMC12060421
- DOI: 10.1186/s40478-025-02013-z
Temporal transcriptomic changes in the THY-Tau22 mouse model of tauopathy display cell type- and sex-specific differences
Abstract
Background: Tauopathies, including Alzheimer's disease (AD) and frontotemporal dementia (FTD), display sex-specific differences in prevalence and progression, but the underlying molecular mechanisms remain unclear. Single-cell transcriptomic analysis of animal models can reveal how AD pathology affects different cell types across sex and age.
Objective: To understand sex-specific and sex-dimorphic transcriptomic changes in different cell types and their age-dependence in the THY-Tau22 mouse model of AD-linked tauopathy.
Methods: We applied single-cell RNA sequencing (scRNA-seq) to cortical tissue from male and female THY-Tau22 and wild-type mice at 17 months of age, when they had prominent tau inclusion pathology, and compared the results with corresponding data previously obtained at 7 months of age. Using differential statistical analysis for individual genes, pathways, and gene regulatory networks, we identified sex-specific, sex-dimorphic, and sex-neutral changes, and looked at how they evolved over age. To validate the most robust findings across distinct mouse models and species, the results were compared with cortical scRNA-seq data from the transgenic hAPP-based Tg2576 mouse model and human AD.
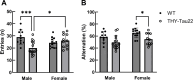
Results: We identified several significant sex-specific and sex-dimorphic differentially expressed genes in neurons, microglia, astrocytes and oligodendrocytes, including both cross-sectional changes and alterations from 7 months to 17 months of age. Key pathways affected in a sex-dependent manner across age included neurotransmitter signaling, RNA processing and splicing, stress response pathways, and protein degradation pathways. In addition, network analysis revealed the AD-associated genes Clu, Mbp, Fos and Junb as relevant regulatory hubs. Analysis of age-dependent changes highlighted genes and pathways associated with inflammatory response (Malat1, Cx3cr1), protein homeostasis (Cst3), and myelin maintenance (Plp1, Cldn11, Mal) that showed consistent sex-dependent changes as the THY-Tau22 mice aged. Multiple genes with established implications in AD, including the long non-coding RNA gene Malat1, displayed concordant sex-specific changes in mouse models and human AD.
Conclusions: This study provides a comprehensive single-cell transcriptomic characterization of sex-linked and age-dependent changes in the THY-Tau22 tauopathy model, revealing new insights into the interplay between age-dependent AD-like pathologies and sex. The identified sex-specific changes and their conservation across models and human AD highlight molecular targets for further preclinical investigation of sex-specific therapeutic strategies in AD.
Keywords: Age differences; Alzheimer's disease; Sex differences; Single-cell RNA sequencing; THY-Tau22 mouse model; Tauopathy; Transcriptomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: The animal experiments adhered to the European FELASA guidelines for animal research and were granted ethical approval by the local Institutional Animal Experimentation Ethics Committee. Furthermore, these experiments were endorsed by the relevant Luxembourg government bodies, including the Ministries of Agriculture and Health. This research did not involve any human subjects. Consent to participate: Does not apply to this study. Competing interests: The authors declare no competing interests.
Figures







References
-
- Schwartz JB, Weintraub S (2021) Treatment for alzheimer Disease—Sex and gender effects need to be explicitly analyzed and reported in clinical trials. JAMA Netw Open 4:e2124386. 10.1001/jamanetworkopen.2021.24386 - PubMed
-
- Dubal DB (2020) Sex difference in Alzheimer’s disease: an updated, balanced and emerging perspective on differing vulnerabilities. Handb Clin Neurol 175:261–273. 10.1016/B978-0-444-64123-6.00018-7 - PubMed
-
- Ossenkoppele R, Lyoo CH, Jester-Broms J, Sudre CH, Cho H, Ryu YH et al (2020) Assessment of demographic, genetic, and imaging variables associated with brain resilience and cognitive resilience to pathological Tau in patients with alzheimer disease. JAMA Neurol 77:632. 10.1001/jamaneurol.2019.5154 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

