Azetidinyl Malachite Green: a superior fluorogen-activating protein probe for live-cell and dynamic SIM imaging
- PMID: 40336989
- PMCID: PMC12053737
- DOI: 10.1039/d5sc01150g
Azetidinyl Malachite Green: a superior fluorogen-activating protein probe for live-cell and dynamic SIM imaging
Abstract
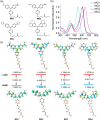
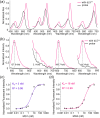
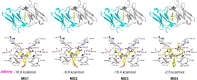
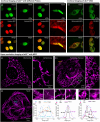
Malachite Green (MG) and its fluorogen-activating protein (FAP) pair are valuable tools for live-cell and super-resolution fluorescence imaging due to their unique near-infrared absorption and signal enhancement. However, the low brightness and photostability of MG have limited its use in dynamic imaging. In this study, we introduce a novel derivative, azetidinly Malachite Green (Aze-MG), which enhances the brightness of the MG-FAP complex by 2.6-fold. This enhancement is achieved by replacing the N,N-dimethylamino group in MG with an azetidine group, which suppresses the twisted intramolecular charge transfer (TICT) effect, leading to improved quantum yield and photostability. Additionally, the reduced binding affinity of Aze-MG for FAP enables a buffering strategy, allowing the reversible exchange of photobleached fluorogens with free fluorogens, thereby ensuring stable fluorescence over time. This combination of improved brightness and buffering capability makes Aze-MG an ideal probe for live-cell and dynamic SIM imaging.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
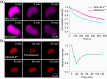
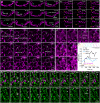
References
-
- Daugird T. A. Shi Y. Holland K. L. Rostamian H. Liu Z. Lavis L. D. Rodriguez J. Strahl B. D. Legant W. R. Correlative single molecule lattice light sheet imaging reveals the dynamic relationship between nucleosomes and the local chromatin environment. Nat. Commun. 2024;15:4178. doi: 10.1038/s41467-024-48562-0. - DOI - PMC - PubMed
-
- Kozma E. Kele P. Fluorogenic probes for super-resolution microscopy. Org. Biomol. Chem. 2019;17:215–233. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

