DNA Methylation Epitypes of Burkitt Lymphoma with Distinct Molecular and Clinical Features
- PMID: 40338627
- PMCID: PMC12209777
- DOI: 10.1158/2643-3230.BCD-24-0240
DNA Methylation Epitypes of Burkitt Lymphoma with Distinct Molecular and Clinical Features
Abstract
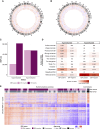
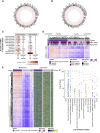
The genetic subtypes of Burkitt lymphoma have been defined, but the role of epigenetics remains to be comprehensively characterized. We searched genomic DNA from 218 patients across four continents for recurrent DNA methylation patterns and their associations with clinical and molecular features. We identified DNA methylation patterns that were not fully explained by the Epstein-Barr virus status or mutation status, leading to two epitypes described here as HypoBL and HyperBL. Each is characterized by distinct genomic and clinical features including global methylation, mutation burden, aberrant somatic hypermutation, and survival outcomes. Methylation, gene expression, and mutational differences between the epitypes support a model in which each arises from a distinct cell of origin. These results, pending validation in external cohorts, point to a refined risk assessment for patients with Burkitt lymphoma who may experience inferior outcomes.
Significance: Burkitt lymphoma can be divided into two epigenetic subtypes (epitypes), each carrying distinct biological, transcriptomic, genomic, and clinical features. Epitype is more strongly associated with clinical and mutational features than the Epstein-Barr virus status or genetic subtype, highlighting an important additional layer of Burkitt lymphoma pathogenesis.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
N. Thomas reports personal fees from Vevo Therapeutics outside the submitted work. C. Casper reports grants from NIH during the conduct of the study. J.M. Gastier-Foster reports grants from NIH during the conduct of the study, as well as other support from Beckman Coulter Biosciences outside the submitted work. A.S. Gerrie reports grants and personal fees from Eli Lilly Canada and BeiGene, personal fees from AstraZeneca, and grants from Janssen outside the submitted work. T.C. Greiner reports other support from Leidos Biomedical Research, Inc., a subcontractor for NIH, during the conduct of the study. C.G. Mullighan reports personal fees from Illumina during the conduct of the study, as well as grants from Pfizer, personal fees from Amgen, and other support from Cyrus outside the submitted work. D.W. Scott reports personal fees from AbbVie, AstraZeneca, Genmab, Kite/Gilead, and Veracyte and grants and personal fees from Roche/Genentech outside the submitted work, as well as a patent for Using Gene Expression to Assign Cell-of-origin Class to Aggressive B-cell Lymphomas issued and licensed to NanoString Technologies and a patent for Using Gene Expression to Identify the Dark Zone Signature in Aggressive B-cell Lymphomas issued. M. Esteller reports grants from Incyte and personal fees from Quimatryx outside the submitted work. No disclosures were reported by the other authors.
Figures





References
MeSH terms
Grants and funding
- 75N91019D00024/CA/NCI NIH HHS/United States
- R35 CA197695/CA/NCI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- 75N91020F00003/National Cancer Institute (NCI)
- CEL007/Fundación Cellex (Cellex Foundation)
- P30 CA008748/CA/NCI NIH HHS/United States
- Canadian Institutes of Health Research (CIHR)
- HHSN261201100063C/CA/NCI NIH HHS/United States
- 1043/Terry Fox Research Institute (TFRI)
- 2021 SGR01494/CERCA Programme/Generalitat de Catalunya
- Burkitt Lymphoma Genome Sequencing Project
- HHSN261201100007I/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- PID2021-125282OB-I00/Spanish Ministry of Science and Innovation
- National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources

