Clade 2.3.4.4b highly pathogenic avian influenza H5N1 viruses: knowns, unknowns, and challenges
- PMID: 40340397
- PMCID: PMC12172449
- DOI: 10.1128/jvi.00424-25
Clade 2.3.4.4b highly pathogenic avian influenza H5N1 viruses: knowns, unknowns, and challenges
Abstract
Since 2020, the clade 2.3.4.4b highly pathogenic avian influenza (HPAI) H5N1 viruses have caused unprecedented outbreaks in wild birds and domestic poultry globally, resulting in significant ecological damage and economic losses due to the disease and enforced stamp-out control. In addition to the avian hosts, the H5N1 viruses have expanded their host range to infect many mammalian species, potentially increasing the zoonotic risk. Here, we review the current knowns and unknowns of clade 2.3.4.4b HPAI H5N1 viruses, and we highlight common challenges in prevention. By integrating our knowledge of viral evolution and ecology, we aim to identify discrepancies and knowledge gaps for a more comprehensive understanding of the virus. Ultimately, this review will serve as a theoretical foundation for researchers involved in related avian influenza virus studies, aiding in improved control and prevention of H5N1 viruses.
Keywords: Clade 2.3.4.4b; H5N1; evolution; highly pathogenic avian influenza; pathogenicity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

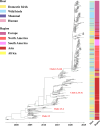

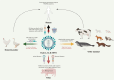
References
Publication types
MeSH terms
Grants and funding
- 32461120064,32473060/National Natural Science Foundation of China
- 2024A1515013151/the National Natural Science Foundation of Guangdong Province
- 2025A04J5445/Guangzhou Basic and Applied Basic Research Project
- 2024/Young Scholars of Yangtze River Scholar Professor Program
- 2024/Young Pearl River Scholar of "Guangdong Special Support Plan"
LinkOut - more resources
Full Text Sources
Medical

