Data Interoperability and Harmonization in Cardiovascular Genomic and Precision Medicine
- PMID: 40340425
- PMCID: PMC12173165
- DOI: 10.1161/CIRCGEN.124.004624
Data Interoperability and Harmonization in Cardiovascular Genomic and Precision Medicine
Abstract
Despite advances in cardiovascular care and improved outcomes, fragmented healthcare systems, nonequitable access to health care, and nonuniform and unbiased collection and access to healthcare data have exacerbated disparities in healthcare provision and further delayed the technological-enabled implementation of precision medicine. Precision medicine relies on a foundation of accurate and valid omics and phenomics that can be harnessed at scale from electronic health records. Big data approaches in noncardiovascular healthcare domains have helped improve efficiency and expedite the development of novel therapeutics; therefore, applying such an approach to cardiovascular precision medicine is an opportunity to further advance the field. Several endeavors, including the American Heart Association Precision Medicine platform and public-private partnerships (such as BigData@Heart in Europe), as well as cloud-based platforms, such as Terra used for the National Institutes of Health All of Us, are attempting to temporally and ontologically harmonize data. This state-of-the-art review summarizes best practices used in cardiovascular genomic and precision medicine and provides recommendations for systems' requirements that could enhance and accelerate the integration of these platforms.
Keywords: big data; electronic health records; natural language processing; phenomics; translational research, biomedical.
Conflict of interest statement
All authors declare no conflicts of interest, no relevant relationships to disclose, and no funding received from any companies or organizations related to the subject of this article.
Figures

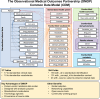
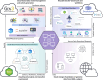


Similar articles
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Defining disease severity in atopic dermatitis and psoriasis for the application to biomarker research: an interdisciplinary perspective.Br J Dermatol. 2024 Jun 20;191(1):14-23. doi: 10.1093/bjd/ljae080. Br J Dermatol. 2024. PMID: 38419411 Free PMC article. Review.
-
SNMMI Procedure Standard/EANM Practice Guideline for Brain [18F]FDG PET Imaging, Version 2.0.J Nucl Med. 2025 Jun 6;66(Suppl 2):S45-S60. doi: 10.2967/jnumed.124.268754. J Nucl Med. 2025. PMID: 39419552
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
Surveillance for Violent Deaths - National Violent Death Reporting System, 50 States, the District of Columbia, and Puerto Rico, 2022.MMWR Surveill Summ. 2025 Jun 12;74(5):1-42. doi: 10.15585/mmwr.ss7405a1. MMWR Surveill Summ. 2025. PMID: 40493548 Free PMC article.
Cited by
-
Machine learning based, subject-specific, gender and race independent, non-invasive estimation of the arterial blood pressure.NPJ Cardiovasc Health. 2025;2(1):41. doi: 10.1038/s44325-025-00075-5. Epub 2025 Aug 1. NPJ Cardiovasc Health. 2025. PMID: 40757190 Free PMC article.
References
-
- Armoundas AA, Narayan SM, Arnett DK, Spector-Bagdady K, Bennett DA, Celi LA, Friedman PA, Gollob MH, Hall JL, Kwitek AE, et al. ; on behalf of the American Heart Association Institute for Precision Cardiovascular Medicine; Council on Cardiovascular and Stroke Nursing; Council on Lifelong Congenital Heart Disease and Heart Health in the Young; Council on Cardiovascular Radiology and Intervention; Council on Hypertension; Council on the Kidney in Cardiovascular Disease; and Stroke Council. Use of artificial intelligence in improving outcomes in heart disease: a scientific statement from the American Heart Association. Circulation. 2024;149:e1028. doi: 10.1161/CIR.0000000000001201 - PMC - PubMed
-
- NIH. NIH broadens genomic data-sharing policy. Cancer Discov. 2014;4:OF4. doi: 10.1158/2159-8290 - PubMed
-
- Bota P, Thambiraj G, Bollepalli SC, Armoundas AA. Artificial intelligence algorithms in cardiovascular medicine: an attainable promise to improve patient outcomes or an inaccessible investment? Curr Cardiol Rep. 2024;26:1477–1485. doi: 10.1007/s11886-024-02146-y - PubMed
-
- Donnelly WJ. Viewpoint: patient-centered medical care requires a patient-centered medical record. Acad Med. 2005;80:33–38. doi: 10.1097/00001888-200501000-00009 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

