Genome content reorganization in the non-model ciliate Chilodonella uncinata: insights into nuclear architecture, DNA content, and chromosome fragmentation during macronuclear development
- PMID: 40340440
- PMCID: PMC12188737
- DOI: 10.1128/msphere.00075-25
Genome content reorganization in the non-model ciliate Chilodonella uncinata: insights into nuclear architecture, DNA content, and chromosome fragmentation during macronuclear development
Abstract
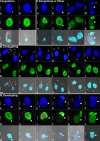
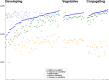
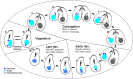
Ciliates are a model lineage for studies of genome architecture given their unusual genome structures. All ciliates have both somatic macronuclei (MAC) and germline micronuclei (MIC), both of which develop from a zygotic nucleus following sex (i.e., conjugation). Nuclear developmental stages are not well documented among non-model ciliates, including Chilodonella uncinata (class Phyllopharyngea), the focus of our work. Here, we characterize nuclear architecture and genome dynamics in C. uncinata by combining 4',6-diamidino-2-phenylindole (DAPI) staining and fluorescence in situ hybridization (FISH) techniques with confocal microscopy. We developed a telomere probe for staining, which alongside DAPI allows for the identification of fragmented somatic chromosomes among the total DNA in the nuclei. We quantify both total DNA and telomere-bound signals from more than 250 nuclei sampled from 116 individual cells, and analyze changes in DNA content and nuclear architecture across Chilodonella's nuclear life cycle. Specifically, we find that MAC developmental stages in the ciliate C. uncinata are different from those reported from other ciliate species. These data provide insights into nuclear dynamics during development and enrich our understanding of genome evolution in non-model ciliates.
Importance: Ciliates are a clade of diverse single-celled eukaryotic microorganisms that contain at least one somatic macronucleus (MAC) and germline micronucleus (MIC) within each cell/organism. Ciliates rely on complex genome rearrangements to generate somatic genomes from a zygotic nucleus. However, the development of somatic nuclei has only been documented for a few model ciliate genera, including Paramecium, Tetrahymena, and Oxytricha. Here, we study the MAC developmental process in the non-model ciliate, C. uncinata. We analyze both total DNA and the generation of gene-sized somatic chromosomes using a laser scanning confocal microscope to describe C. uncinata's nuclear life cycle. We show that DNA content changes dramatically during their life cycle and in a manner that differs from previous studies on model ciliates. Our study expands knowledge of genome dynamics in ciliates and among eukaryotes more broadly.
Keywords: DAPI; DNA content; FISH; chromosomes; ciliate; confocal imaging; genes; genome dynamics; macronuclei; nuclear cycle; single-celled microorganisms; telomere.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Update of
-
Genome content in the non-model ciliate Chilodonella uncinata: insights into nuclear architecture, gene-sized chromosomes among the total DNA in their somatic macronuclei during their development.bioRxiv [Preprint]. 2024 Nov 14:2024.11.13.623465. doi: 10.1101/2024.11.13.623465. bioRxiv. 2024. Update in: mSphere. 2025 Jun 25;10(6):e0007525. doi: 10.1128/msphere.00075-25. PMID: 39605396 Free PMC article. Updated. Preprint.
Similar articles
-
Genome content in the non-model ciliate Chilodonella uncinata: insights into nuclear architecture, gene-sized chromosomes among the total DNA in their somatic macronuclei during their development.bioRxiv [Preprint]. 2024 Nov 14:2024.11.13.623465. doi: 10.1101/2024.11.13.623465. bioRxiv. 2024. Update in: mSphere. 2025 Jun 25;10(6):e0007525. doi: 10.1128/msphere.00075-25. PMID: 39605396 Free PMC article. Updated. Preprint.
-
Exploration of the Germline Genome of the Ciliate Chilodonella uncinata through Single-Cell Omics (Transcriptomics and Genomics).mBio. 2018 Jan 9;9(1):e01836-17. doi: 10.1128/mBio.01836-17. mBio. 2018. PMID: 29317511 Free PMC article.
-
Macronuclear development in ciliates, with a focus on nuclear architecture.J Eukaryot Microbiol. 2022 Sep;69(5):e12898. doi: 10.1111/jeu.12898. Epub 2022 Mar 16. J Eukaryot Microbiol. 2022. PMID: 35178799 Free PMC article. Review.
-
Alternative processing of scrambled genes generates protein diversity in the ciliate Chilodonella uncinata.J Exp Zool B Mol Dev Evol. 2010 Sep 15;314(6):480-8. doi: 10.1002/jez.b.21354. J Exp Zool B Mol Dev Evol. 2010. PMID: 20700892 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

