Targeted next-generation sequencing characterization of respiratory pathogens in children with acute respiratory infection
- PMID: 40340627
- PMCID: PMC12060355
- DOI: 10.1186/s12879-025-11051-w
Targeted next-generation sequencing characterization of respiratory pathogens in children with acute respiratory infection
Abstract
Background: Acute respiratory infections (ARIs) pose a significant global health burden, particularly affecting infants and young children with high morbidity and mortality rates. Targeted next-generation sequencing (tNGS) has emerged as a rapid and cost-effective diagnostic tool capable of identifying a broad range of respiratory tract infections.
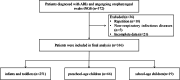
Methods: Oropharyngeal swabs and sputum samples were collected from patients and subjected to tNGS and sputum culture, respectively, for diagnosing ARIs. A retrospective analysis was conducted on clinical data to explore the clinical diagnosis and therapeutic application of tNGS.
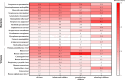
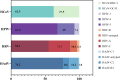
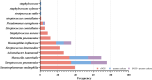
Results: This study included 336 pediatric patients with confirmed ARIs. tNGS detected 38 potential pathogens, comprising 25 species (15 bacteria and 10 viruses) and 13 viral subtypes. The overall microbial detection rate using tNGS was 100%. The leading bacterial pathogens identified were Streptococcus pneumoniae (36.0%), Stenotrophomonas maltophilia (30.4%), Streptococcus intermedius (29.5%), Moraxella catarrhalis (27.1%), and Hemophilus influenzae (20.2%). The predominant viral pathogens included human adenovirus (31.3%), human rhinovirus (26.5%), human parainfluenza virus (25.0%), cytomegalovirus (19.0%), and human bocavirus (11.0%). Among the 94 patients who underwent simultaneous sputum culture and Gram staining, tNGS exhibited a superior detection rate compared to sputum culture (100% vs. 53.2%). Among the 50 patients with concordant positive results for both tNGS and sputum culture, 80% (40/50) demonstrated full or partial agreement. Additionally, tNGS revealed age-specific heterogeneity in pathogen distribution across different age groups.
Conclusion: Traditional diagnostic methods often fall short of meeting the diagnostic demands of ARIs. This study underscores the potential of tNGS in oropharyngeal swabs for enhancing pathogen detection, thereby improving the diagnosis, treatment, and prevention of ARIs.
Importance: This study represents the first investigation utilizing oropharyngeal swabs for tNGS in diagnosing and treating ARIs. By analyzing surveillance data from a local hospital's patients with ARIs, we have identified the spectrum of bacterial and viral pathogens and explored demographic differences among patients. These findings underscore the potential of tNGS in ARI surveillance, diagnosis, pathogen detection, and prevention.
Keywords: Acute respiratory infections; Diagnosis; Pathogen spectrum; Pharyngeal swabs; Targeted next-generation sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the Ethics Committee of Binyang Women and Children’s Hospital and was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients’ parents or their legal representatives. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests. Conflict of interest: The authors have no competing interests to declare. Clinical trial number: Not applicable.
Figures
Similar articles
-
Interpretation of pathogenicity and clinical features of multiple pathogens in pediatric lower respiratory tract infections by tNGS RPTM analysis.Eur J Clin Microbiol Infect Dis. 2025 Jun;44(6):1313-1324. doi: 10.1007/s10096-025-05094-9. Epub 2025 Mar 14. Eur J Clin Microbiol Infect Dis. 2025. PMID: 40085381
-
Clinical diagnostic value of throat swabs in pediatric acute lower respiratory tract infections using targeted next-generation sequencing.BMC Pediatr. 2025 Mar 21;25(1):224. doi: 10.1186/s12887-024-05380-z. BMC Pediatr. 2025. PMID: 40114075 Free PMC article.
-
Comparative performance evaluation of FilmArray RP 2.1 and targeted next-generation sequencing in upper respiratory tract infections.Front Cell Infect Microbiol. 2025 Jul 24;15:1610445. doi: 10.3389/fcimb.2025.1610445. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40778290 Free PMC article.
-
Rethinking Paediatric Respiratory Infections: The Role of Mixed Pathogen Infections.Rev Med Virol. 2025 Mar;35(2):e70021. doi: 10.1002/rmv.70021. Rev Med Virol. 2025. PMID: 40000823 Review.
-
Detecting Respiratory Pathogens for Diagnosing Lower Respiratory Tract Infections at the Point of Care: Challenges and Opportunities.Biosensors (Basel). 2025 Feb 20;15(3):129. doi: 10.3390/bios15030129. Biosensors (Basel). 2025. PMID: 40136926 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources

