Circulating immune cells exhibit distinct traits linked to metastatic burden in breast cancer
- PMID: 40340807
- PMCID: PMC12063295
- DOI: 10.1186/s13058-025-01982-2
Circulating immune cells exhibit distinct traits linked to metastatic burden in breast cancer
Abstract
Background: Circulating immune cells play a crucial role in the anti-tumour immune response, yet the systemic immune system in metastatic breast cancers is not fully characterised. Investigating the cellular and molecular changes in peripheral blood mononuclear cells (PBMCs) from breast cancer patients could elucidate the role of circulating immune cells in metastasis and aid in identifying biomarkers for disease burden and progression.
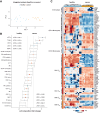
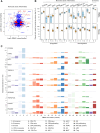
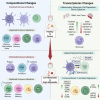
Methods: In this study, we characterised the systemic immune landscape associated with varying levels of metastatic burden by analysing the single-cell transcriptomes of PBMCs from breast cancer patients and healthy controls. Our research focused on identifying changes in immune cell composition, transcriptional programs, and immune-cell communication networks linked to metastatic burden. Additionally, we compared these PBMC features onto a single-cell atlas of primary breast tumours to study corresponding traits in tumour-infiltrating immune cells.
Results: In metastatic breast cancer, PBMCs exhibit a significant downregulation of the adaptive immune system and a decreased number and activity of unconventional T cells, such as γδ T cells. Additionally, metastatic burden is associated with impaired cell communication pathways involved in immunomodulatory functions. We also identified a gene signature derived from myeloid cells shared between tumour immune infiltrates and circulating immune cells in breast cancer patients.
Conclusions: Our study provides a comprehensive single-cell molecular profile of the peripheral immune system in breast cancer, offering a valuable resource for understanding metastatic disease in terms of tumour burden. By identifying immune traits linked to metastasis, we have unveiled potential new biomarkers of metastatic disease.
Keywords: Breast cancer; Metastasis; PBMCs; Single-cell RNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: Human breast tissues and blood samples were obtained from consenting patients through the Austin Health, a tertiary cancer hospital. Human Ethics approval was obtained from the Austin Human Research Ethics Committee (HREC/66494/Austin-2020). Clinicopathological characterisation, including the site of metastasis, is provided in supplementary tables (Tables S1, S2 and S3). Consent for publications: Not applicable Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Comprehensive single-cell chromatin and transcriptomic profiling of peripheral immune cells in nonsegmental vitiligo.Br J Dermatol. 2025 Jun 20;193(1):115-124. doi: 10.1093/bjd/ljaf041. Br J Dermatol. 2025. PMID: 39888372
-
Systematic screening of metabolic pathways to identify two breast cancer subtypes with divergent immune characteristics.Sci Rep. 2025 Jul 1;15(1):20996. doi: 10.1038/s41598-025-05179-7. Sci Rep. 2025. PMID: 40594370 Free PMC article.
-
CD177, MYBL2, and RRM2 Are Potential Biomarkers for Musculoskeletal Infections.Clin Orthop Relat Res. 2025 Jun 1;483(6):1062-1071. doi: 10.1097/CORR.0000000000003402. Epub 2025 Feb 6. Clin Orthop Relat Res. 2025. PMID: 39915095
-
Breast surgery for metastatic breast cancer.Cochrane Database Syst Rev. 2018 Mar 15;3(3):CD011276. doi: 10.1002/14651858.CD011276.pub2. Cochrane Database Syst Rev. 2018. PMID: 29542106 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

