Precision enhancement of CAR-NK cells through non-viral engineering and highly multiplexed base editing
- PMID: 40341025
- PMCID: PMC12067936
- DOI: 10.1136/jitc-2024-009560
Precision enhancement of CAR-NK cells through non-viral engineering and highly multiplexed base editing
Abstract
Background: Natural killer (NK) cells' unique ability to kill transformed cells expressing stress ligands or lacking major histocompatibility complexes (MHC) has prompted their development for immunotherapy. However, NK cells have demonstrated only moderate responses against cancer in clinical trials.
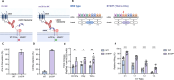
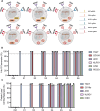
Methods: Advanced genome engineering may thus be used to unlock their full potential. Multiplex genome editing with CRISPR/Cas9 base editors (BEs) has been used to enhance T cell function and has already entered clinical trials but has not been reported in human NK cells. Here, we report the first application of BE in primary NK cells to achieve both loss-of-function and gain-of-function mutations.
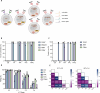
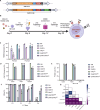
Results: We observed highly efficient single and multiplex base editing, resulting in significantly enhanced NK cell function in vitro and in vivo. Next, we combined multiplex BE with non-viral TcBuster transposon-based integration to generate interleukin-15 armored CD19 chimeric antigen receptor (CAR)-NK cells with significantly improved functionality in a highly suppressive model of Burkitt's lymphoma both in vitro and in vivo.
Conclusions: The use of concomitant non-viral transposon engineering with multiplex base editing thus represents a highly versatile and efficient platform to generate CAR-NK products for cell-based immunotherapy and affords the flexibility to tailor multiple gene edits to maximize the effectiveness of the therapy for the cancer type being treated.
Keywords: Chimeric antigen receptor - CAR; Gene therapy; Hematologic Malignancies; Immunotherapy; Natural killer - NK.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: MW, EJP, MGK, BM and BRW have filed patents covering the methods and approaches outlined in this work. All other authors declare they have no competing interests.
Figures



Update of
-
Precision Enhancement of CAR-NK Cells through Non-Viral Engineering and Highly Multiplexed Base Editing.bioRxiv [Preprint]. 2024 Mar 8:2024.03.05.582637. doi: 10.1101/2024.03.05.582637. bioRxiv. 2024. Update in: J Immunother Cancer. 2025 May 7;13(5):e009560. doi: 10.1136/jitc-2024-009560. PMID: 38496503 Free PMC article. Updated. Preprint.
Similar articles
-
Precision Enhancement of CAR-NK Cells through Non-Viral Engineering and Highly Multiplexed Base Editing.bioRxiv [Preprint]. 2024 Mar 8:2024.03.05.582637. doi: 10.1101/2024.03.05.582637. bioRxiv. 2024. Update in: J Immunother Cancer. 2025 May 7;13(5):e009560. doi: 10.1136/jitc-2024-009560. PMID: 38496503 Free PMC article. Updated. Preprint.
-
Concurrent transposon engineering and CRISPR/Cas9 genome editing of primary CLL-1 chimeric antigen receptor-natural killer cells.Cytotherapy. 2022 Nov;24(11):1087-1094. doi: 10.1016/j.jcyt.2022.07.008. Epub 2022 Aug 29. Cytotherapy. 2022. PMID: 36050244
-
Revolutionising Cancer Immunotherapy: Advancements and Prospects in Non-Viral CAR-NK Cell Engineering.Cell Prolif. 2025 Apr;58(4):e13791. doi: 10.1111/cpr.13791. Epub 2024 Dec 27. Cell Prolif. 2025. PMID: 39731215 Free PMC article. Review.
-
Engineering the next generation of CAR-NK immunotherapies.Int J Hematol. 2021 Nov;114(5):554-571. doi: 10.1007/s12185-021-03209-4. Epub 2021 Aug 28. Int J Hematol. 2021. PMID: 34453686 Free PMC article. Review.
-
Use of Cell and Genome Modification Technologies to Generate Improved "Off-the-Shelf" CAR T and CAR NK Cells.Front Immunol. 2020 Aug 7;11:1965. doi: 10.3389/fimmu.2020.01965. eCollection 2020. Front Immunol. 2020. PMID: 32903482 Free PMC article. Review.
Cited by
-
A Singular Base Editing Platform for Polyfunctional Multiplex Engineering of Immune Cells.bioRxiv [Preprint]. 2025 Jul 16:2025.07.11.664404. doi: 10.1101/2025.07.11.664404. bioRxiv. 2025. PMID: 40791471 Free PMC article. Preprint.
-
Overcoming Immune Barriers in Allogeneic CAR-NK Therapy: From Multiplex Gene Editing to AI-Driven Precision Design.Biomolecules. 2025 Jun 26;15(7):935. doi: 10.3390/biom15070935. Biomolecules. 2025. PMID: 40723807 Free PMC article. Review.
References
-
- Miller JS, Lanier LL. Natural Killer Cells in Cancer Immunotherapy. Annu Rev Cancer Biol. 2019;3:77–103. doi: 10.1146/annurev-cancerbio-030518-055653. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
