In silico identification of gene targets to enhance C12 fatty acid production in Escherichia coli
- PMID: 40341454
- PMCID: PMC12062052
- DOI: 10.1007/s00253-025-13501-6
In silico identification of gene targets to enhance C12 fatty acid production in Escherichia coli
Abstract
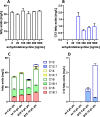
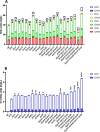
The global interest in fatty acids is steadily rising due to their wealth of industrial potential ranging from cosmetics to biofuels. Unfortunately, certain fatty acids, such as monounsaturated lauric acid with a carbon atom chain length of twelve (C12 fatty acids), cannot be produced cost and energy-efficiently using conventional methods. Biosynthesis using microorganisms can overcome this drawback. However, rewiring a microbe's metabolome for increased production remains challenging. To overcome this, sophisticated genome-wide metabolic network models have become available. These models predict the effect of genetic perturbations on the metabolism, thereby serving as a guide for metabolic pathways optimization. In this work, we used constraint-based modeling in combination with the algorithm Optknock to identify gene deletions in Escherichia coli that improve C12 fatty acid production. Nine gene targets were identified that, when deleted, were predicted to increase C12 fatty acid titers. Targets play a role in anaplerotic reactions, amino acid synthesis, carbon metabolism, and cofactor-balancing. Subsequently, we constructed the corresponding (combinatorial) deletion mutants to validate the in silico predictions in vivo. Our highest producer (ΔmaeB Δndk ΔpykA) reaches a titer of 6.7 mg/L, corresponding to a 7.5-fold increase in C12 fatty acid production. This study demonstrates that model-guided metabolic engineering is a useful tool to improve C12 fatty acid production. KEY POINTS: •Escherichia coli as a promising biofactory for unsaturated C12 fatty acids. •Optknock to identify non-obvious gene deletions for increased C12 fatty acids. •7.5-fold higher C12 fatty acid production achieved by deleting maeB, ndk, and pykA.
Keywords: Escherichia coli; C12 fatty acids; Lauric acid; Model-guided metabolic engineering; Oleochemicals; Optknock.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical statement: This article does not contain any studies with human participants or animals performed by any of the authors. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C-metabolic flux analysis.Biotechnol Bioeng. 2014 Mar;111(3):575-85. doi: 10.1002/bit.25124. Biotechnol Bioeng. 2014. PMID: 24122357 Free PMC article.
-
Constraint-based modeling of heterologous pathways: application and experimental demonstration for overproduction of fatty acids in Escherichia coli.Biotechnol Bioeng. 2014 Oct;111(10):2056-66. doi: 10.1002/bit.25261. Epub 2014 Jun 16. Biotechnol Bioeng. 2014. PMID: 24838438
-
Enhanced production of branched-chain fatty acids by replacing β-ketoacyl-(acyl-carrier-protein) synthase III (FabH).Biotechnol Bioeng. 2015 Aug;112(8):1613-22. doi: 10.1002/bit.25583. Epub 2015 Apr 17. Biotechnol Bioeng. 2015. PMID: 25788017
-
Biosynthesis of ω-hydroxy fatty acids and related chemicals from natural fatty acids by recombinant Escherichia coli.Appl Microbiol Biotechnol. 2019 Jan;103(1):191-199. doi: 10.1007/s00253-018-9503-6. Epub 2018 Nov 11. Appl Microbiol Biotechnol. 2019. PMID: 30417307 Review.
-
Metabolic engineering of microorganisms for the production of structurally diverse esters.Appl Microbiol Biotechnol. 2017 Apr;101(8):3043-3053. doi: 10.1007/s00253-017-8179-7. Epub 2017 Mar 8. Appl Microbiol Biotechnol. 2017. PMID: 28275821 Review.
References
-
- Bosco ML, Varrica D, Dongarrà G (2005) Case study: Inorganic pollutants associated with particulate matter from an area near a petrochemical plant. Environ Res 99:18–30. 10.1016/j.envres.2004.09.011 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

